Publications - Supplemental material
Please find below supplemental material corresponding to publications of our group. Currently, we list 133 supplements.
If you have problems accessing electronic information, please let us know:
You may use this URL to cite or link to us.
©NOTICE: All documents are copyrighted by the authors; If you would like to use all or a portion of any paper, please contact the author.
This supplement is also available at http://www.bioinf.uni-leipzig.de/publications/supplements/07-001You may use this URL to cite or link to us.
BIOINF 07-001: Computational RNomics of Drosophilids
Dominic Rose, Jörg Hackermüller, Stefan Washietl, Sven Findeiß, Kristin Reiche, Jana Hertel, Peter F. Stadler, Sonja J. Prohaska
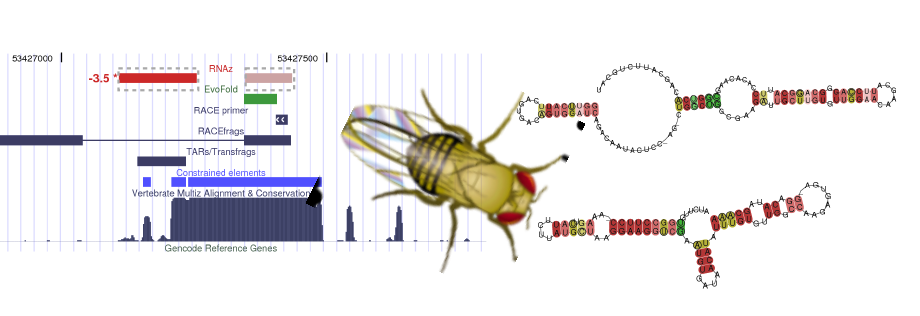

The analysis is based on the genomic data of the CAF1 freeze which contains D.mel release 4.3, see
http://rana.lbl.gov/drosophila/assemblies.html or
ftp://ftp.flybase.net/genomes/aaa/
- D. melanogaster ncRNA candidate sequences (p>0.5, fasta):
all (6.9 Mb) [2L (1.3 Mb) - 2R (1.1 Mb) - 3L (1.5 Mb) - 3R (1.7 Kb) - 4 (34 Kb) - X (1.5 Mb)] - D. melanogaster ncRNA candidate coordinates (p>0.5, bed):
all [2L - 2R - 3L - 3R - 4 - X] - D. melanogaster ncRNA candidate annotation (p>0.5):
2L - 2R - 3L - 3R - 4 - X - Browsing the RNAz hits:
2L (7824 loci), 2R (6646 loci), 3L (8765 loci), 3R (10351 loci), 4 (196 loci), X (8700 loci)The custom annotation field of the above html tables comprises:
- Overlap to known RNAs of reference genome ('known=')
- Overlap to known CDS elements of reference genome ('knownCDS=')
- A blast search against the Rfam ('Rfam=')
- A blast search against the Noncode ('Noncode=')
- A blast search against the miRBase ('miRBase=')
- A blast search against the Noncode ('ncRNAdb=')
- A blast search against the Flybase ('Flybase=')
- Applying tRNAscan ('tRNAscan=')
- Applying RNAmicro ('RNAmicro=')
- Complete WPGMA clustering tree in newick format: [here]
It can be viewed with: http://pbil.univ-lyon1.fr/software/njplot.html - PDF containing additional figures and tables















