RNAdesign: Computational Design Multistable RNA Structures
Using our novel tool, RNAdesign, we make it possible to efficienctly design multi-stable RNA sequences. RNAdesign combines a graph-coloring based approach with a sampling method that allows us to explore the space of candidate sequences for the given structural designs very efficiently.
You can read about RNAdesign here: ‘Höner zu Siederdissen et al. (2013)’.
The sources also available via hackage and github. Additional scripts, and example structural targets are on github as well.
The images below show three (very small) target structures and the ViennaRNA dotplot (far right) for the designed sequence.
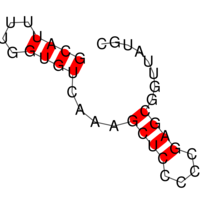
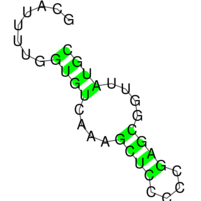
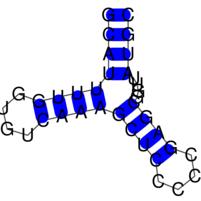

Christian Höner zu Siederdissen, Stefan Hammer, Ingrid Abfalter, Ivo L. Hofacker, Christoph Flamm, and Peter F. Stadler. 2013. Computational design of RNAs with complex energy landscapes. Biopolymers 99, no. 12. 99: 1124–36. doi:10.1002/bip.22337.
README
(included from project)RNAdesign
The RNAdesign program solves the multi-target RNA sequence design problem. You can give one or more structural targets for which a single compatible sequence is designed.
PAPER
Christian Hoener zu Siederdissen, Stefan Hammer, Ingrid Abfalter, Ivo L. Hofacker, Christoph Flamm, Peter F. Stadler. Computational Design of RNAs with Complex Energy Landscapes. 2013. Biopolymers. 99, no. 12. 99. 1124–36. http://dx.doi.org/10.1002/bip.22337.
Contact
choener@tbi.univie.ac.at
HOW TO USE RNAdesign
RNAdesign designs RNA sequences given one or more structural targets. The program offers a variety of optimization functions that each can be used to optimize candidate sequence towards a certain goal, say, minimal ensemble defect or small energetic distance to another target structure.
RNAdesign input
Structural targets are given via stdin, preferably via an input file. Below is a the small tri-stable from our paper, which you should then pipe to RNAdesign: “echo tri-stable.dat | RNAdesign”
“cat tri-stable.dat:”
# a tri-stable example target. (optional comment) ((((….))))….((((….))))…….. ……..((((….((((….))))….)))) ((((((((….))))((((….))))….)))) # below follows a simple (and optional) sequence constraint. CKNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNB
The input may contain many comments lines, starting with a hash “#” and at most one sequence constraint line. All of these lines are optional, except of course for the structural constraints.
Optimization functions
Depending on the actual design you are looking for, you’ll want to modify the optimization function. Below, the different options available are detailed. By giving a complex “–optfun”, many different design goals can be tried.
A good optimization goal is (as an example for three targets):
–optfun “eos(1)+eos(2)+eos(3) - 3 * gibbs + 1 * ((eos(1)-eos(2))^2 + (eos(1)-eos(3))^2 + (eos(2)-eos(3))^2)”
This way, the sequence produces close-to-mfe foldings with the targets (left) and the targets are close together in terms of energy. (1 * ) scales the two terms according to user choice.
binary, combining:
- / :: the four basic operations ^ :: (^) generalized power function
binary, apply function to many targets:
sum max min :: run function over set of targets: sum(eos,1,2) or sum(eos,all)
unary, apply to single target:
eos :: energy of a structure: eos(1) ed :: ensemble defect of a structure: ed(3) partc :: constrained partition function: partc(1).
You probably want to use partc in conjunction with eos, where eos is modified by a small constant: “0.1 * eos(1) + partc(1)”. eos guides the optimizer to the first viable sequence, after which the constrained partition function becomes active.
nullary, constant for the current sequence:
Ged :: global, weighted ensemble defect: Ged gibbs :: gibbs free energy of sequence mfe :: minimum free energy of sequence
special:
logMN :: requires four parameters logMN(0.2,0.3,0.3,0.2) penalizes according to given mono-nucleotide distribution in order of ACGU

|

|

|

|
|
| Binaries | Stable Sources | git | Bugtracker | Build Status |
Installation from sources via stack:
This software can be compiled and installed using stack in a small number of steps. Note that some packages only provide a library and no executable.- Prepare a directory for the software: mkdir ~/haskell/ cd ~/haskell (If you choose another name, replace accordingly below)
- Install stack itself. Binaries are available here. Copy the binaries into the ~/haskell directory.
- Download the RNAdesign software: ~/haskell/stack unpack RNAdesign ls This unpacks the newest version of RNAdesign, the version itself being referred to as -VERSION, which you should see via ls.
- Compile: cd ~/haskell/RNAdesign-VERSION ~/haskell/stack build
- Execute (you can find the executable names, if any, in the cabal as executable build targets): ~/haskell/stack exec EXECUTABLE
- Optionally, install any executables into ~/haskell/bin/. This simplifies actually running the programs later on. mkdir -p ~/haskell/bin ~/haskell/stack install --local-bin-path ~/haskell/bin