Chromatin Computation: Epigenetic Inheritance as a Pattern Reconstruction Problem
References:
Arnold, C, Stadler, P.F., Prohaska, S.J. 2013. Chromatin Computation: Epigenetic Inheritance as a Pattern Reconstruction Problem. Journal of Theoretical Biology 336(7): 61-74. [PDF]
Description
Eukaryotic histones carry a diverse set of specific chemical
modifications that accumulate over the life-time of a cell and have a
crucial impact on the cell state in general and the transcriptional
program in particular. Replication constitutes a dramatic disruption
of the chromatin states that effectively amounts to partial erasure of
stored information. To preserve its epigenetic state the cell
reconstructs (at least part of) the histone modifications by means of
processes that are still very poorly understood. A plausible hypothesis
is that the different combinations of reader and writer domains in
histone-modifying enzymes implement local rewriting rules that are
capable of ``recomputing'' the desired parental modification patterns on
the basis of the partial information contained in that half of the
nucleosomes that predate replication.
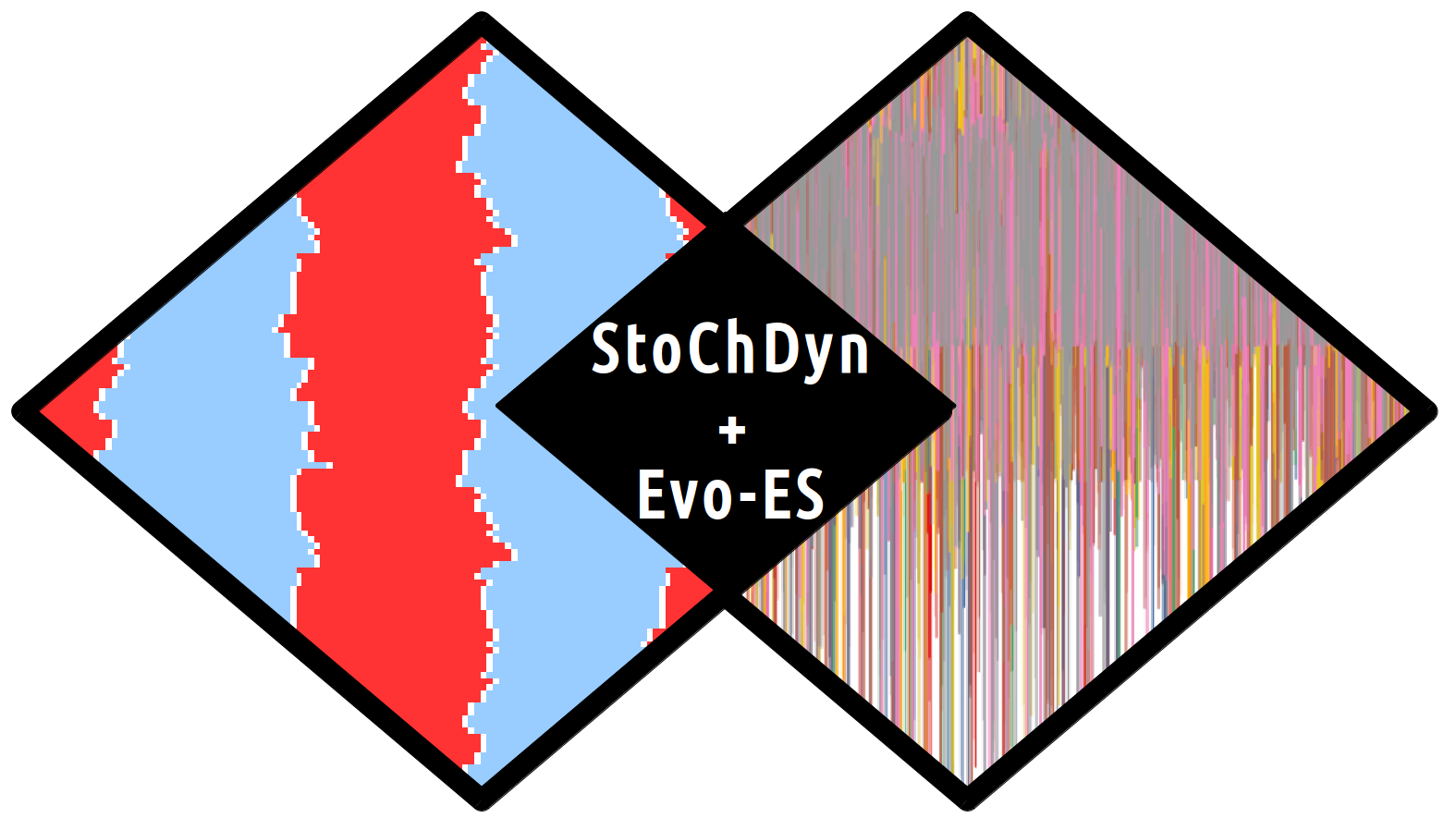
To test whether such a mechanism is theoretically feasible, we have
developed a flexible stochastic simulation system (available at http://www.bioinf.uni-leipzig.de/Software/StoChDyn) for studying the
dynamics of histone modification states. The implementation is based on
Gillespie's approach, i.e., it models the master equation of a detailed
chemical model. It is efficient enough to use an
evolutionary algorithm to find patterns across multiple cell divisions with high accuracy.
We found that it is easy to evolve a system of enzymes that
can maintain a particular chromatin state roughly stable, even without explicit
boundary elements separating differentially modified chromatin
domains. However, the success of this task depends on several
previously unanticipated factors, such as the length of the initial
state, the specific pattern that should be maintained, the time between
replications, and chemical parameters such as enzymatic binding and dissociation
rates. All these factors also influence
the accumulation of errors in the wake of cell divisions.