The 10kTrees Project
Reference:
Arnold, C., Matthews, L. J., Nunn, C. L. 2010.
The 10kTrees Website: A New Online Resource for Primate Phylogeny. Evolutionary Anthropology 19:114-118. [PDF]
CHRISTIAN ARNOLD, PhD


Arnold, C., Matthews, L. J., Nunn, C. L. 2010.
The 10kTrees Website: A New Online Resource for Primate Phylogeny. Evolutionary Anthropology 19:114-118. [PDF]
The 10kTrees Website is a new web resource for conducting
phylogenetic comparative studies of primates. The comparative method
plays a central role in efforts to uncover the adaptive basis for
primate behaviors, morphological traits and cognitive abilities. Using
new phylogeny-based methods, it is now possible to incorporate
evolutionary history directly into comparative research.
The true phylogeny of a group of organisms is never known with
certainty, and phylogenetic relationships should be continually
reassessed as new data become available. This last fact recommends
against the continued use of older phylogenies, as better data are now
available. Furthermore, when conducting a comparative test, different
trees can produce different results during comparative analysis, which
argues against conditioning comparative tests on a single hypothesis of
evolutionary relationships when that hypothesis is legitimately
uncertain. A major development in recent phylogenetics research involves
the use of statistical methods that control for phylogenetic uncertainty
through application of Bayesian phylogenetics. We follow these
recommendations by providing a set of trees that are sampled in
proportion to their posterior probabilities using Markov chain Monte
Carlo (MCMC). This allows researchers to run analyses on an entire set
of trees rather than using a single tree; thus, results are then no
longer conditioned on a single tree being correct.
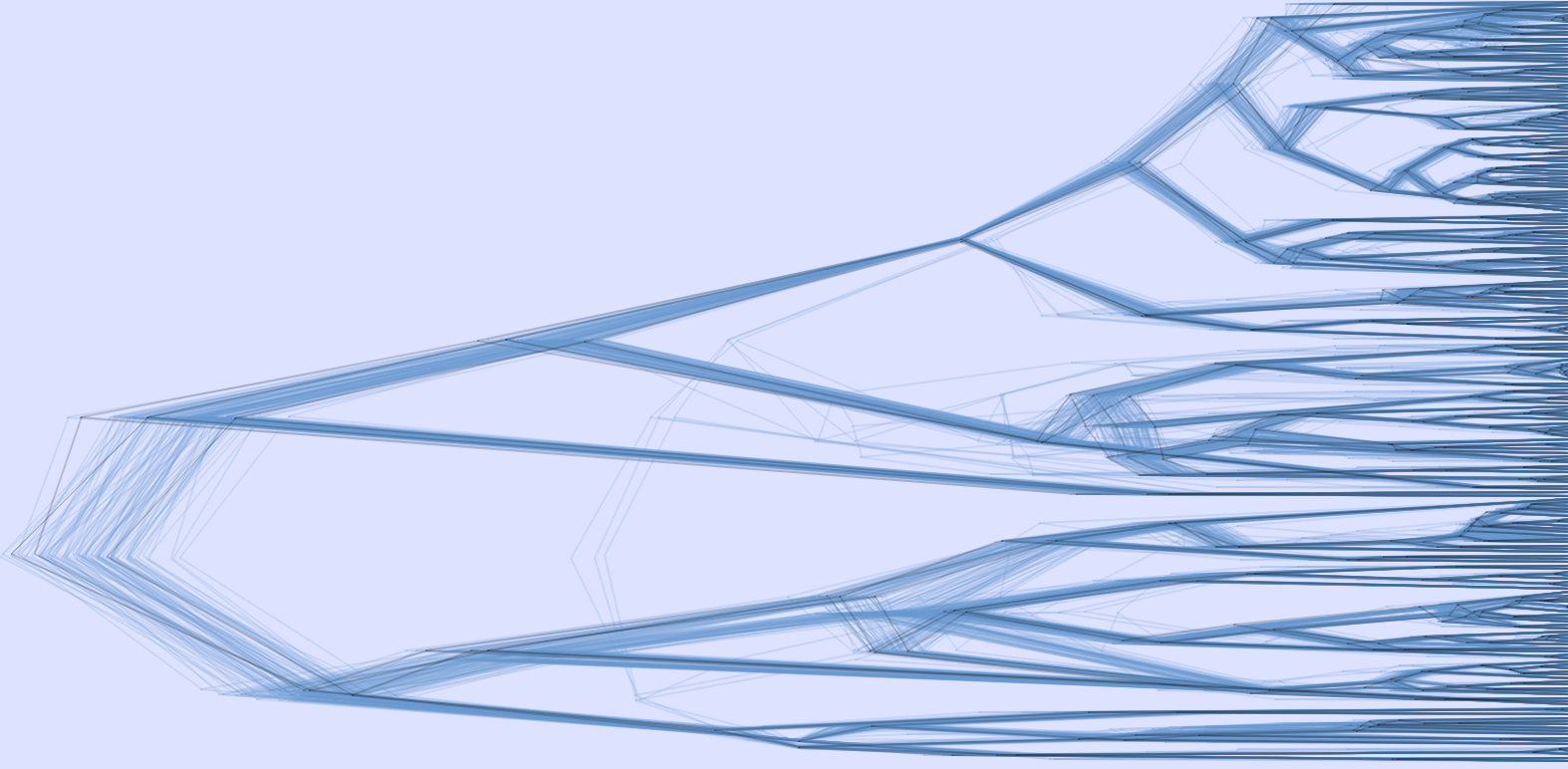
Using the 10kTrees Website, users can download up to at least 10,000
primate phylogenies (with branch lengths) sampled from a Bayesian tree
inference. Moreover, we designed the website so that it can be easily
updated as new versions of the phylogeny become available. For example,
Version 3 of the dataset is already available, which includes over 300 species and additional genes as compared to Version 1 and Version 2. We also expect
that the website itself will evolve to provide more tools for primate
comparative biology.
What do you do once you have your trees? A number of programs are well
suited to analyzing a set of trees in a comparative context. BayesTraits
provides a way to investigate correlated evolution and reconstruct
ancestral states, and it does so very readily across multiple trees,
including through Bayesian approaches. In addition, the PDAP module of
Mesquite can be used to calculate independent contrasts and reconstruct
ancestral states on a tree-by-tree basis. Mesquite can be used to view
the trees, as can the program FigTree. We also formatted the output
files so that they can be read by the APE package of R, which opens up a
wide array of analysis options.
For more information, see the 10kTrees Website.