
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,769,220 – 6,769,331 |
| Length | 111 |
| Max. P | 1.000000 |

| Location | 6,769,220 – 6,769,331 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.15 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -19.47 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.72 |
| SVM decision value | 8.05 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6769220 111 + 22407834 --GAUAC--GAUACGAAGUGCAAGGAUCAAAUACUAUCAAGGAUCAAUGAUUAAUAAAUUAACAUUUCAUUUACAAUUAUAACU----GAUCCUUGACACAAUUUUCGCCGAGCAAAGA --.....--.........(((..((...((((....((((((((((((((((..(((((.........))))).))))))...)----)))))))))....))))...))..))).... ( -20.70) >DroEre_CAF1 38252 87 + 1 --------AGAUUCGAGGUGCUCGGAUCAAAUGGUGUCAAGGAUAA--------UAUC------------CUGCAGUGCUAAGU----GAUCCUUGACACAAUUUUCUCCGAGCAAAGA --------....((....((((((((..((((.(((((((((((..--------....------------..((...)).....----.))))))))))).))))..))))))))..)) ( -33.74) >DroYak_CAF1 38350 99 + 1 ACGAUACGAGAUUCGUGGUGCAAGGAUCAAAUAGUAUCAAGGAUAG--------UACA------------UUAUAAUUAAAAGAAUUGUAUCCUUGAUACAAUUUUCUCCGAGCAAAGA ((((........))))..(((..(((..((((.((((((((((...--------....------------.(((((((.....))))))))))))))))).))))..)))..))).... ( -27.10) >consensus __GAUAC_AGAUUCGAGGUGCAAGGAUCAAAUAGUAUCAAGGAUAA________UAAA____________UUACAAUUAUAAGU____GAUCCUUGACACAAUUUUCUCCGAGCAAAGA ..................(((..(((..((((.((((((((((((...........................................)))))))))))).))))..)))..))).... (-19.47 = -20.36 + 0.89)

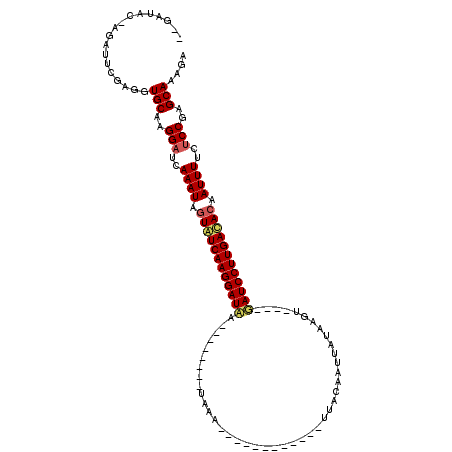
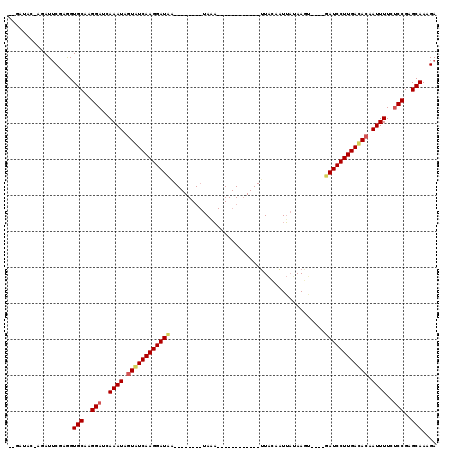
| Location | 6,769,220 – 6,769,331 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.15 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.38 |
| Structure conservation index | 0.73 |
| SVM decision value | 7.45 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6769220 111 - 22407834 UCUUUGCUCGGCGAAAAUUGUGUCAAGGAUC----AGUUAUAAUUGUAAAUGAAAUGUUAAUUUAUUAAUCAUUGAUCCUUGAUAGUAUUUGAUCCUUGCACUUCGUAUC--GUAUC-- ....(((..((...((((..(((((((((((----(((.((....((((((.........))))))..)).))))))))))))))..))))...))..))).........--.....-- ( -27.10) >DroEre_CAF1 38252 87 - 1 UCUUUGCUCGGAGAAAAUUGUGUCAAGGAUC----ACUUAGCACUGCAG------------GAUA--------UUAUCCUUGACACCAUUUGAUCCGAGCACCUCGAAUCU-------- ....((((((((..((((.(((((((((((.----.(((.((...))))------------)...--------..))))))))))).))))..))))))))..........-------- ( -34.70) >DroYak_CAF1 38350 99 - 1 UCUUUGCUCGGAGAAAAUUGUAUCAAGGAUACAAUUCUUUUAAUUAUAA------------UGUA--------CUAUCCUUGAUACUAUUUGAUCCUUGCACCACGAAUCUCGUAUCGU ....(((..(((..((((.((((((((((((..................------------....--------.)))))))))))).))))..)))..)))..(((.....)))..... ( -26.80) >consensus UCUUUGCUCGGAGAAAAUUGUGUCAAGGAUC____ACUUAUAAUUGUAA____________UAUA________UUAUCCUUGAUACUAUUUGAUCCUUGCACCUCGAAUCU_GUAUC__ ....(((..(((..((((.((((((((((((...........................................)))))))))))).))))..)))..))).................. (-21.43 = -21.76 + 0.34)

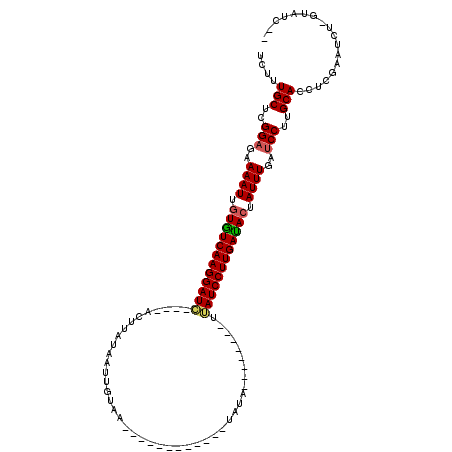
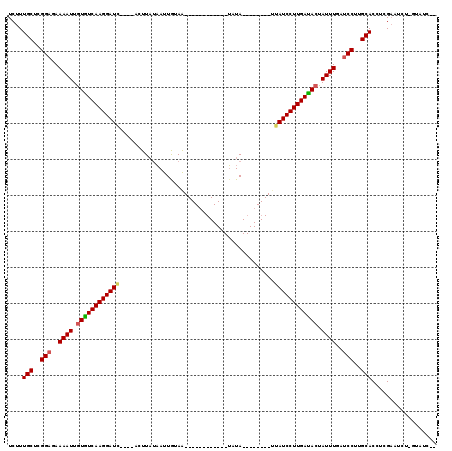
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:38 2006