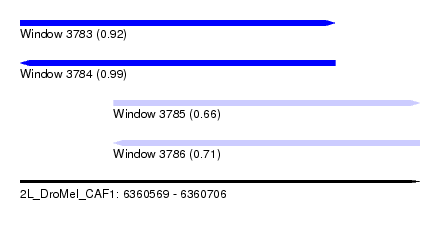
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,360,569 – 6,360,706 |
| Length | 137 |
| Max. P | 0.991941 |

| Location | 6,360,569 – 6,360,677 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.54 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -10.88 |
| Energy contribution | -13.44 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6360569 108 + 22407834 -----GCAAAUACGGG---CGAGUUAUAGGAUCCGUGCUCUCGGUGGGGUAACCG-CCAAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGUUCUGUUCCCAGCCGCCCC-CAUUUGG -----.(((((..(((---((.(((((((((((.(((((...(((((.....)))-)).)))))((.(((((....))))).)).....))))))))....)))))))).-.))))). ( -36.30) >DroSec_CAF1 8308 108 + 1 -----GCAAAUACGGG---CGAGUUAUAGGAUCCGUGCUCUCGGUGGGGUAACCG-CCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGUUCGGUUCCCAGCCGCCCC-CAUUUGG -----.(((((..(((---((.(((...(((.(((.((.((((((((.....)))-))))).))((.(((((....))))).)).........))).))).)))))))).-.))))). ( -41.30) >DroEre_CAF1 8132 101 + 1 -----GCAAAUACGGG---CGAGUUAUGGGAUCCGUGCUCUCGGUGGGGUAACCG-CCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGU-----UCCCAGCCGCCCC-CAUU--G -----........(((---((.((..(((((.(((.((.((((((((.....)))-))))).))((.(((((....))))).))....))).-----)))))))))))).-....--. ( -42.50) >DroWil_CAF1 23025 97 + 1 UAUUUGAGAGUGAAAG---UGAGGAGCCAAAUCCA--CCCGCAGUU-GGUAACCAAACACAUGCGGUAAUUUACCUAAUUUUCAACGC---UUC---UCCC----CCUCA-CAU---- .........((((..(---.(((((((........--.(((((.((-((...)))).....))))).((((.....))))......))---)))---)).)----..)))-)..---- ( -23.00) >DroYak_CAF1 8473 104 + 1 --------AAAGCGGGAUAGGAGUUAUUGGAUCCCUGCUCUCGGUGGGGUAACCG-CCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGC-----UCGCAGCCGCCCCCCAUUUGG --------.....(((....(((..(((((((....((.((((((((.....)))-))))).))........))))))).))).....((((-----.....))))...)))...... ( -34.60) >consensus _____GCAAAUACGGG___CGAGUUAUAGGAUCCGUGCUCUCGGUGGGGUAACCG_CCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGUUC___UCCCAGCCGCCCC_CAUUUGG .............(((....((((............)))).((((.(((.......((((.((.((.(((((....))))).)).)))))).......))).)))))))......... (-10.88 = -13.44 + 2.56)

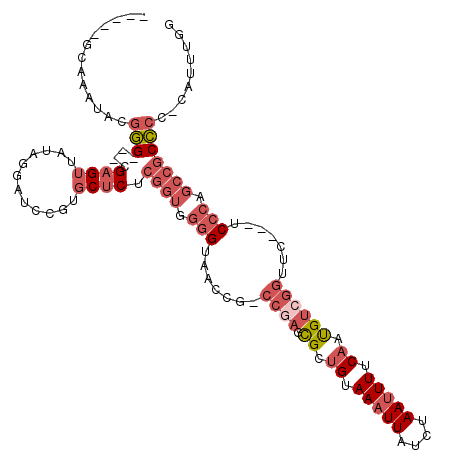
| Location | 6,360,569 – 6,360,677 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.54 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.16 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6360569 108 - 22407834 CCAAAUG-GGGGCGGCUGGGAACAGAACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUUGG-CGGUUACCCCACCGAGAGCACGGAUCCUAUAACUCG---CCCGUAUUUGC----- .((((((-.(((((((((....)))..(((....((.(((((....))))).))((.(((((-.((.....)).))))).)).)))..........)))---))).)))))).----- ( -38.00) >DroSec_CAF1 8308 108 - 1 CCAAAUG-GGGGCGGCUGGGAACCGAACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGG-CGGUUACCCCACCGAGAGCACGGAUCCUAUAACUCG---CCCGUAUUUGC----- .((((((-.((((((.(((((.(((.........((.(((((....))))).))((.(((((-.((.....)).))))).)).))).)))))....)))---))).)))))).----- ( -42.20) >DroEre_CAF1 8132 101 - 1 C--AAUG-GGGGCGGCUGGGA-----ACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGG-CGGUUACCCCACCGAGAGCACGGAUCCCAUAACUCG---CCCGUAUUUGC----- .--((((-.((((((.(((((-----.(((....((.(((((....))))).))((.(((((-.((.....)).))))).)).))).)))))....)))---))).))))...----- ( -40.70) >DroWil_CAF1 23025 97 - 1 ----AUG-UGAGG----GGGA---GAA---GCGUUGAAAAUUAGGUAAAUUACCGCAUGUGUUUGGUUACC-AACUGCGGG--UGGAUUUGGCUCCUCA---CUUUCACUCUCAAAUA ----..(-(((((----(.((---(.(---((..(....).....((((((.(((((.((...(((...))-)))))))).--..))))))))).))).---)))))))......... ( -26.00) >DroYak_CAF1 8473 104 - 1 CCAAAUGGGGGGCGGCUGCGA-----GCCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGG-CGGUUACCCCACCGAGAGCAGGGAUCCAAUAACUCCUAUCCCGCUUU-------- .....(((((((..(((((((-----((.....(((.(((((....))))).))).)))).)-))))..)))).)))(((((.(((((............))))))))))-------- ( -36.40) >consensus CCAAAUG_GGGGCGGCUGGGA___GAACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGG_CGGUUACCCCACCGAGAGCACGGAUCCUAUAACUCG___CCCGUAUUUGC_____ .........(((..((((..................................))))((((...((((......)))).))))....................)))............. (-12.36 = -12.16 + -0.20)

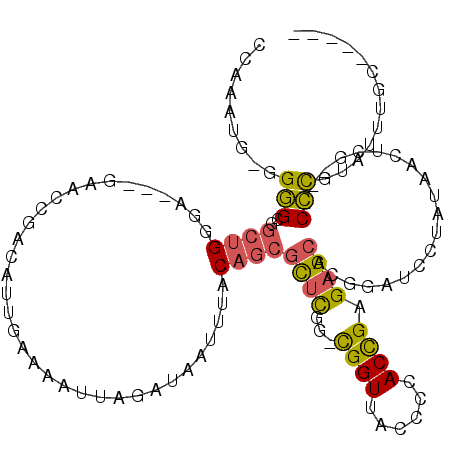
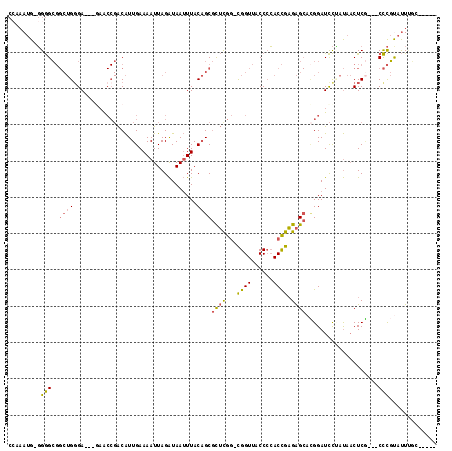
| Location | 6,360,601 – 6,360,706 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -24.24 |
| Energy contribution | -25.55 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6360601 105 + 22407834 UCGGUGGGGUAACCGCCAAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGUUCUGUUCCCAGCCGCCCC-CAUUUGG-GAUACUGAUUGGAUUUCAGUAUUGUUCCU .((((((((.(((((....(((.((.(((((....))))).)).))))))))....))))).)))..((-(....))-)(((((((.......)))))))....... ( -28.00) >DroSec_CAF1 8340 101 + 1 UCGGUGGGGUAACCGCCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGUUCGGUUCCCAGCCGCCCC-CAUUUGG-GG----GAUUGGAUUUCAGUAUUGUUCCU .((((.(((.(((((((((.((.((.(((((....))))).)).))))))..)))))))).))))((((-(....))-))----)...(((............))). ( -36.70) >DroEre_CAF1 8164 99 + 1 UCGGUGGGGUAACCGCCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGU-----UCCCAGCCGCCCC-CAUU--GGGGUGCUGAUUGGAUUUCAGUACUGUUCCU (((((((.....)))))))(..(((......((((((((..(......(((-----.....)))(((((-....--))))))..))))))))..)))..)....... ( -33.10) >DroYak_CAF1 8505 102 + 1 UCGGUGGGGUAACCGCCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGC-----UCGCAGCCGCCCCCCAUUUGGGGGUACUGAUUGGAUUUCAGUAUUGUUCCU (((((((.....)))))))..((((......((((((((.........(((-----.....)))((((((.....))))))...))))))))..))))......... ( -37.30) >consensus UCGGUGGGGUAACCGCCGAGCGCUGUAAAUUAUCUAAUUUUCAAUGUCGGU_____UCCCAGCCGCCCC_CAUUUGG_GGUACUGAUUGGAUUUCAGUAUUGUUCCU (((((((.....)))))))..((((......((((((((........(((((........)))))((((......)))).....))))))))..))))......... (-24.24 = -25.55 + 1.31)

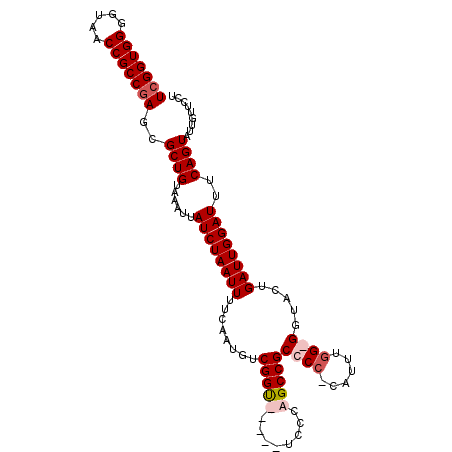
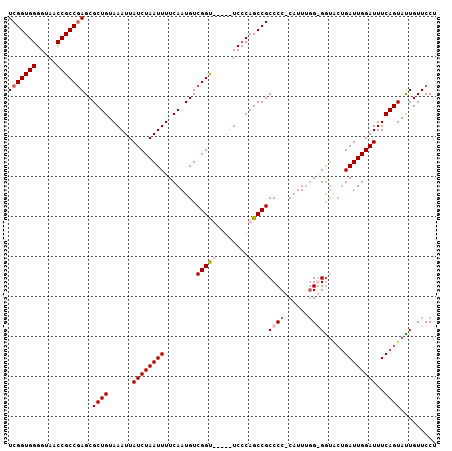
| Location | 6,360,601 – 6,360,706 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -22.14 |
| Energy contribution | -23.57 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6360601 105 - 22407834 AGGAACAAUACUGAAAUCCAAUCAGUAUC-CCAAAUG-GGGGCGGCUGGGAACAGAACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUUGGCGGUUACCCCACCGA .((....(((((((.......))))))).-.....((-(((....(((....)))(((((.(((((.(((((....))))).)))....)).))))).))))))).. ( -27.80) >DroSec_CAF1 8340 101 - 1 AGGAACAAUACUGAAAUCCAAUC----CC-CCAAAUG-GGGGCGGCUGGGAACCGAACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGGCGGUUACCCCACCGA .(((............)))...(----((-((....)-))))(((..((((((((..(((((.(((.(((((....))))).))).).))))))))).)))..))). ( -33.80) >DroEre_CAF1 8164 99 - 1 AGGAACAGUACUGAAAUCCAAUCAGCACCCC--AAUG-GGGGCGGCUGGGA-----ACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGGCGGUUACCCCACCGA .((....((((((...(((...((((.((((--....-))))..)))))))-----.(((((.(((.(((((....))))).))).).))))))).)))....)).. ( -26.40) >DroYak_CAF1 8505 102 - 1 AGGAACAAUACUGAAAUCCAAUCAGUACCCCCAAAUGGGGGGCGGCUGCGA-----GCCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGGCGGUUACCCCACCGA .((.....((((((.......))))))...))...(((((((..(((((((-----((.....(((.(((((....))))).))).)))).)))))..)))).))). ( -32.80) >consensus AGGAACAAUACUGAAAUCCAAUCAGUACC_CCAAAUG_GGGGCGGCUGGGA_____ACCGACAUUGAAAAUUAGAUAAUUUACAGCGCUCGGCGGUUACCCCACCGA .((....(((((((.......)))))))..........((((..((((.........(((((.(((.(((((....))))).))).).))))))))..)))).)).. (-22.14 = -23.57 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:54 2006