| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,285,159 – 6,285,258 |
| Length | 99 |
| Max. P | 0.998424 |

| Location | 6,285,159 – 6,285,258 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.19 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.86 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
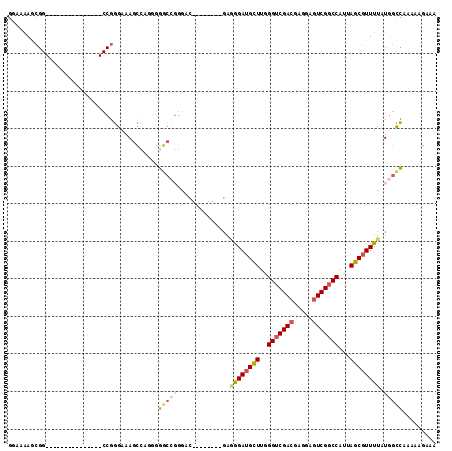
>2L_DroMel_CAF1 6285159 99 + 22407834 GAAAAAGCGG---------------CCGGGAGAGCCAGAGGGCCGGGACGUGGCCGGGAGGGAUGCUUGGGUCGACGAGGAGUCGGCCAUUAGCGUUUUAUGGCCAAAAAGAAA .......(((---------------((.((....))....))))).....((((((...((((((((..(((((((.....)))))))...)))))))).))))))........ ( -42.00) >DroSec_CAF1 121419 93 + 1 GGAAAAACGG---------------CCGGGAAAGCCA-GGGGCCGGGAUGG-----GGAGGGAUGCUUGGGUCGACGAGGAGUCGGCCAUUAGCGUUUUAUGGCCAAAAAGAAA .......(((---------------((.((....)).-..)))))...(((-----...((((((((..(((((((.....)))))))...))))))))....)))........ ( -34.50) >DroSim_CAF1 136113 99 + 1 GGAAAAGCGG---------------CCGGGAAAGCCACGGGGCCGGGACGGGGCCGGGAGGGAUGCUUGGGUCGACGAGGAGUCGGCCAUUAGCGUUUUAUGGCCAAAAAGAAA .......(((---------------((.((....))....)))))......(((((...((((((((..(((((((.....)))))))...)))))))).)))))......... ( -41.10) >DroEre_CAF1 121094 106 + 1 GGAAGAGCGGGAGAGACCGGGAAACCCGGGAAAGCCAGGGGGCCGGGCC--------GAAGGAUGCUUGGGUCGACGAGCAGUCGGCCAUUAGCGUUUUAUGGCCAAAAAGAAA .......(((......)))((...(((.((....)).)))..)).((((--------(.((((((((..(((((((.....)))))))...)))))))).)))))......... ( -46.10) >DroYak_CAF1 134427 80 + 1 GGAAAAGCGG---------------CCGGGAAAACGAAGGG-------------------AGAUGCUUGGGUCGACGAGGAGUCGGCCAUUAGCGUUUUAUGGCCAAAAAGAAA ........((---------------(((......(....)(-------------------(((((((..(((((((.....)))))))...)))))))).)))))......... ( -31.50) >DroAna_CAF1 72971 82 + 1 GCAAGA---------------------GUGAAAGU-AGGGUGCA--AAC--------GAAAGAGGUUUCGGCCGACGUGGAUUCGACCAUUAGCGUUUUAUGGUAAAAAGGAAA ......---------------------......((-((..(((.--..(--------....).((((((.((....)).))...))))....)))..))))............. ( -13.90) >consensus GGAAAAGCGG_______________CCGGGAAAGCCAGGGGGCCGGGAC________GAGGGAUGCUUGGGUCGACGAGGAGUCGGCCAUUAGCGUUUUAUGGCCAAAAAGAAA ........................................(((((..............((((((((..(((((((.....)))))))...)))))))).)))))......... (-20.11 = -20.86 + 0.75)


| Location | 6,285,159 – 6,285,258 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.19 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -9.63 |
| Energy contribution | -9.68 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.765930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
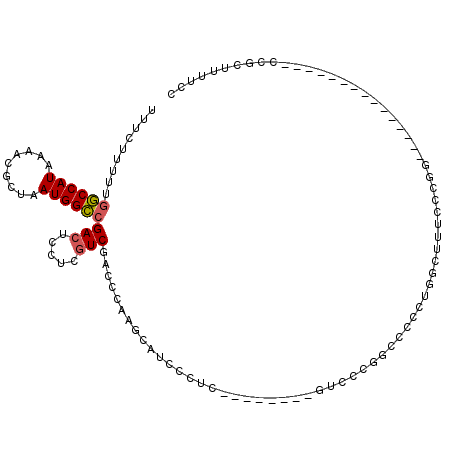
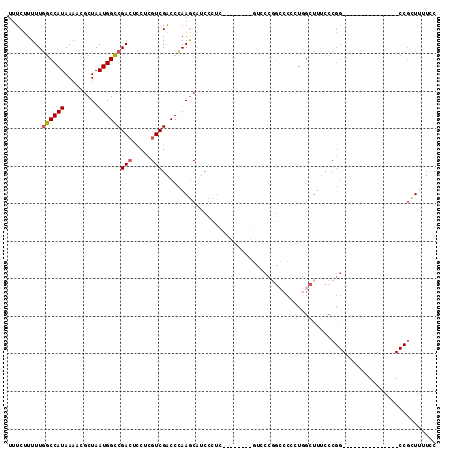
>2L_DroMel_CAF1 6285159 99 - 22407834 UUUCUUUUUGGCCAUAAAACGCUAAUGGCCGACUCCUCGUCGACCCAAGCAUCCCUCCCGGCCACGUCCCGGCCCUCUGGCUCUCCCGG---------------CCGCUUUUUC .........((((.......(((...((.((((.....)))).))..)))...((....((((.......))))....)).......))---------------))........ ( -27.30) >DroSec_CAF1 121419 93 - 1 UUUCUUUUUGGCCAUAAAACGCUAAUGGCCGACUCCUCGUCGACCCAAGCAUCCCUCC-----CCAUCCCGGCCCC-UGGCUUUCCCGG---------------CCGUUUUUCC .........((((.......(((...((.((((.....)))).))..)))........-----.......((((..-.)))).....))---------------))........ ( -22.30) >DroSim_CAF1 136113 99 - 1 UUUCUUUUUGGCCAUAAAACGCUAAUGGCCGACUCCUCGUCGACCCAAGCAUCCCUCCCGGCCCCGUCCCGGCCCCGUGGCUUUCCCGG---------------CCGCUUUUCC .........((((.......(((...((.((((.....)))).))..)))........((....))....))))..((((((.....))---------------))))...... ( -27.70) >DroEre_CAF1 121094 106 - 1 UUUCUUUUUGGCCAUAAAACGCUAAUGGCCGACUGCUCGUCGACCCAAGCAUCCUUC--------GGCCCGGCCCCCUGGCUUUCCCGGGUUUCCCGGUCUCUCCCGCUCUUCC .........(((((......(((...((((((.((((.(......).))))....))--------)))).)))....)))))...(((((...)))))................ ( -28.30) >DroYak_CAF1 134427 80 - 1 UUUCUUUUUGGCCAUAAAACGCUAAUGGCCGACUCCUCGUCGACCCAAGCAUCU-------------------CCCUUCGUUUUCCCGG---------------CCGCUUUUCC .........((((..((((((((...((.((((.....)))).))..)))....-------------------......)))))...))---------------))........ ( -16.90) >DroAna_CAF1 72971 82 - 1 UUUCCUUUUUACCAUAAAACGCUAAUGGUCGAAUCCACGUCGGCCGAAACCUCUUUC--------GUU--UGCACCCU-ACUUUCAC---------------------UCUUGC ....................((.(((((((((.......))))))((((....))))--------)))--.)).....-........---------------------...... ( -11.50) >consensus UUUCUUUUUGGCCAUAAAACGCUAAUGGCCGACUCCUCGUCGACCCAAGCAUCCCUC________GUCCCGGCCCCCUGGCUUUCCCGG_______________CCGCUUUUCC .........((((((.........))))))(((.....)))......................................................................... ( -9.63 = -9.68 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:56 2006