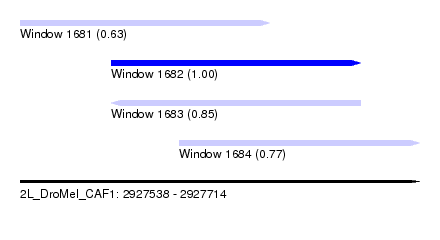
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,927,538 – 2,927,714 |
| Length | 176 |
| Max. P | 0.996193 |

| Location | 2,927,538 – 2,927,648 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -11.81 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | 0.34 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2927538 110 + 22407834 UGCGCCAGCACUACUCAAGCAAUUCAAUACCAACUCAAUAUUGGCUAACCCAAAAAAUGAAAAAA----------AAGAAAAAGAAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUU (((....)))........((((((......(..((((...((((.....))))....(....)..----------...................))))..)........))))))..... ( -11.94) >DroSim_CAF1 46929 103 + 1 UGCGCCAGCACUACUCAAGCAAUUCAAUACCAACUCAAUAUUGGCUAACCCAAAA-------AAA----------AAGCAAAAUAAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUU (((....)))....................(((((......((((...(((....-------...----------......................))))))).....)))))...... ( -11.65) >DroEre_CAF1 46874 108 + 1 UGCGCCAGCAGUACUCAAGCAAUUCAAUACCAACUCAAUAUUGGCUAACCCAAAAAAUUAAAAAAAUACGAAUAAAA------------CAGGAUGAGGGGCCACAAAAAGUUGCAGUUU (((....)))........((((((......(..((((...((((.....))))...............(........------------..)..))))..)........))))))..... ( -11.84) >consensus UGCGCCAGCACUACUCAAGCAAUUCAAUACCAACUCAAUAUUGGCUAACCCAAAAAAU_AAAAAA__________AAG_AAAA_AAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUU (((....)))....................(((((......((((...(((..............................................))))))).....)))))...... (-11.25 = -11.25 + 0.00)

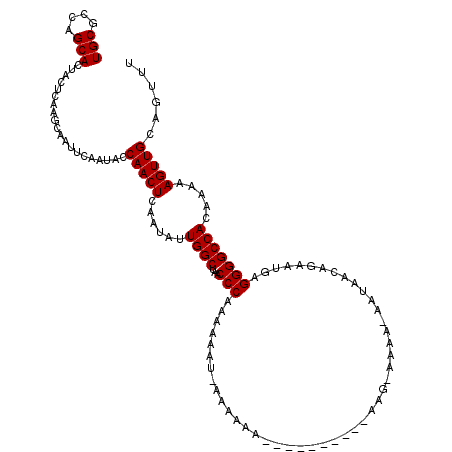
| Location | 2,927,578 – 2,927,688 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -22.80 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2927578 110 + 22407834 UUGGCUAACCCAAAAAAUGAAAAAA----------AAGAAAAAGAAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACAUCAACA ((((((..(((......((......----------..............))......)))......((((((..(.........)..))))))))))))((((((((...))).))))). ( -22.85) >DroSim_CAF1 46969 103 + 1 UUGGCUAACCCAAAA-------AAA----------AAGCAAAAUAAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACAUCAACA ((((((..(((....-------...----------......................)))......((((((..(.........)..))))))))))))((((((((...))).))))). ( -23.05) >DroEre_CAF1 46914 108 + 1 UUGGCUAACCCAAAAAAUUAAAAAAAUACGAAUAAAA------------CAGGAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACGUCAACA ((((((..(((..........................------------........)))......((((((..(.........)..))))))))))))((((((((...))).))))). ( -22.88) >consensus UUGGCUAACCCAAAAAAU_AAAAAA__________AAG_AAAA_AAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACAUCAACA ((((.....))))..............................................(((((...(((((..(.........)..))))))))))..((((((((...))).))))). (-22.80 = -22.80 + 0.00)

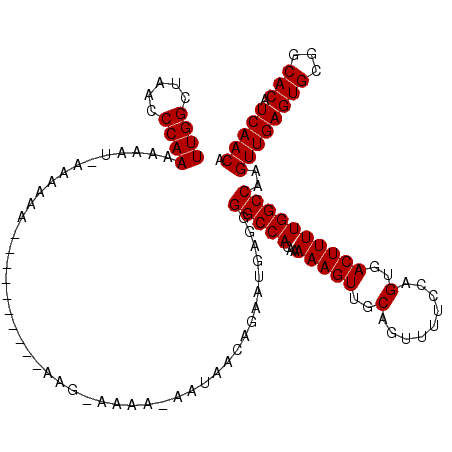
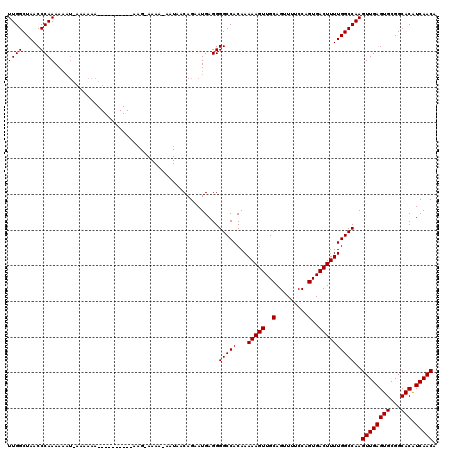
| Location | 2,927,578 – 2,927,688 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -18.15 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | 0.20 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2927578 110 - 22407834 UGUUGAUGUGCCGCACUCAACUUGGCCAAAAGUCACUGGAAAACUGCAACUUUUUGUGGCCCCUCAUUCUGUUAUUCUUUUUCUU----------UUUUUUCAUUUUUUGGGUUAGCCAA .(((((.(((...)))))))).((((.....(((((.(((((........))))))))))(((......................----------..............)))...)))). ( -18.42) >DroSim_CAF1 46969 103 - 1 UGUUGAUGUGCCGCACUCAACUUGGCCAAAAGUCACUGGAAAACUGCAACUUUUUGUGGCCCCUCAUUCUGUUAUUAUUUUGCUU----------UUU-------UUUUGGGUUAGCCAA .(((((.(((...)))))))).((((.((......(..(((((..((((..(...(((((..........))))).)..))))..----------.))-------)))..).)).)))). ( -18.80) >DroEre_CAF1 46914 108 - 1 UGUUGACGUGCCGCACUCAACUUGGCCAAAAGUCACUGGAAAACUGCAACUUUUUGUGGCCCCUCAUCCUG------------UUUUAUUCGUAUUUUUUUAAUUUUUUGGGUUAGCCAA .(((((.(((...)))))))).((((.....(((((.(((((........))))))))))(((........------------..........................)))...)))). ( -18.48) >consensus UGUUGAUGUGCCGCACUCAACUUGGCCAAAAGUCACUGGAAAACUGCAACUUUUUGUGGCCCCUCAUUCUGUUAUU_UUUU_CUU__________UUUUUU_AUUUUUUGGGUUAGCCAA .(((((.(((...)))))))).((((.....(((((.(((((........))))))))))(((..............................................)))...)))). (-18.15 = -18.15 + 0.00)

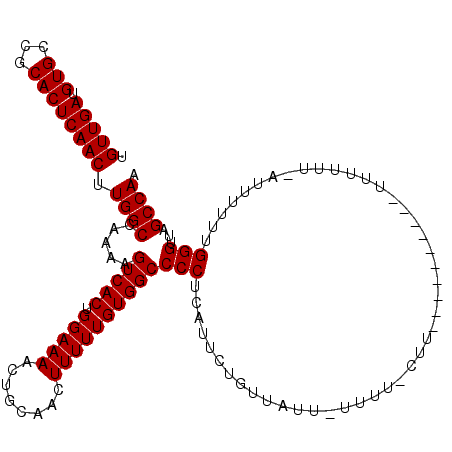
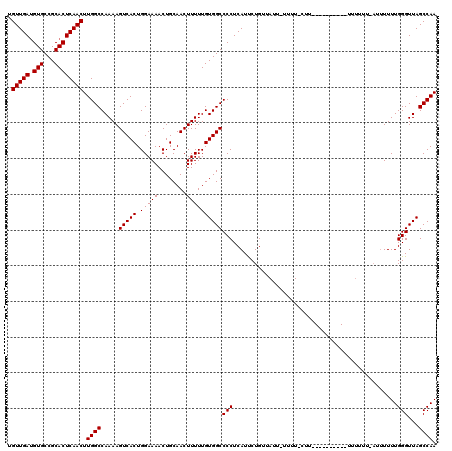
| Location | 2,927,608 – 2,927,714 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -25.14 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2927608 106 + 22407834 AAAGAAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACAUCAACAGGAUUUCAUUUCGCUCUCUC--------UUUCGC (((((....(..((((((((((((...(((((..(.........)..))))))))))..((((((((...))).)))))....)))))))..).....))--------)))... ( -27.40) >DroSim_CAF1 46992 98 + 1 AAAUAAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACAUCAACAGGAUUUCAUCUCUCUCU----------------C ..........(((..(((((((((...(((((..(.........)..))))))))))..((((((((...))).))))).........))))..)))----------------. ( -26.50) >DroEre_CAF1 46951 105 + 1 ---------CAGGAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACGUCAACAGGAUUUCAUUUCGCUCUCUCUCCCUCCCUCUCGC ---------.(((..(((((((((...(((((..(.........)..)))))))))).....(((.((((...(((.....)))......)))).)))....)))))))..... ( -29.60) >consensus AAA_AAUAACAGAAUGAGGGGCCACAAAAAGUUGCAGUUUUCCAGUGACUUUUGGCCAAGUUGAGUGCGGCACAUCAACAGGAUUUCAUUUCGCUCUCUC________U_UCGC ..........(((..(((((((((...(((((..(.........)..))))))))))..((((((((...))).))))).........))))..)))................. (-25.14 = -24.70 + -0.44)

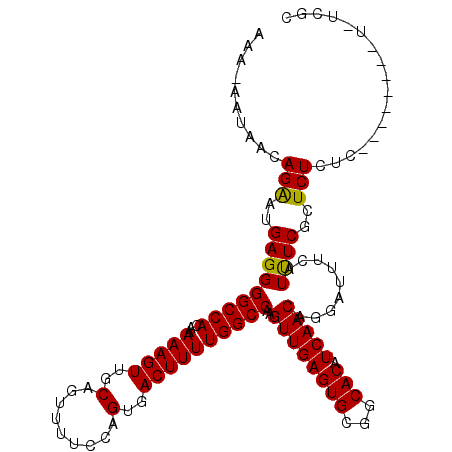
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:58 2006