Genomic and epigenomic co-evolution in follicular lymphomas
 Reference:
Reference:
M Loeffler, M Kreuz, A Haake, D Hasenclever, H Trautmann, C Arnold, K Winter, K Koch, W Klapper, R Scholtysik, M Rosolowski, S Hoffmann, O Ammerpohl, M Szczepanowski, D Herrmann, R Kueppers, C Pott, R Siebert and on behalf of the HaematoSys-Project. 2014.
Genomic and epigenomic co-evolution in follicular lymphomas. Leukemia (16 July 2014) | doi:10.1038/leu.2014.209
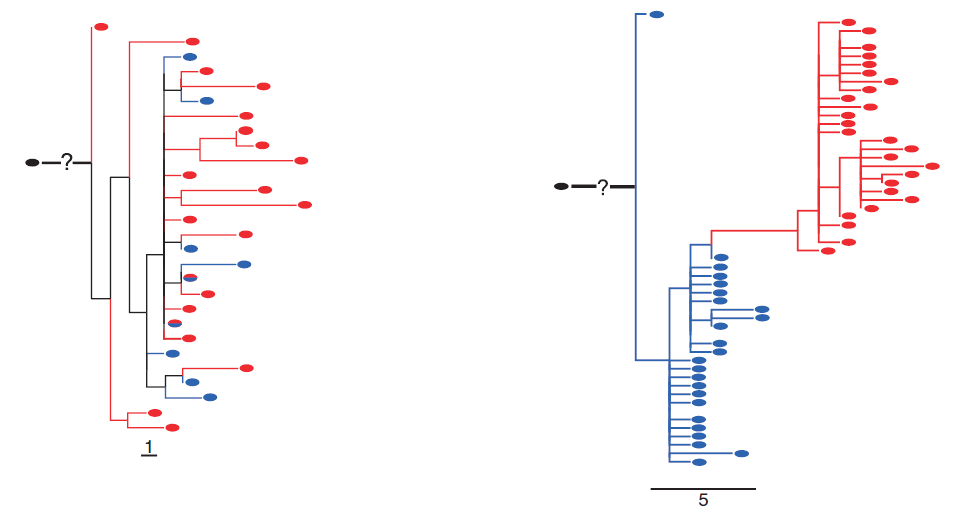
Follicular lymphoma (FL) with a t(14;18) is a B-cell neoplasm clinically characterized by multiple recurrencies. In order to investigate the clonal evolution of this lymphoma, we studied paired primary and relapse tumor samples from 33 patients with recurrent non-transformed t(14;18)-positive FL. We reconstructed phylogenetic trees of the evolution by taking advantage of the activation-induced cytidine deaminase (AID)-mediated somatic hypermutation (SHM) active in the germinal center reaction using sequences of the clonal VHDHJH rearrangements of the immunoglobulin heavy chain (IGH) locus. Mutational analysis of the IGH locus showed evidence for ongoing somatic mutation and for counter-selection of mutations affecting the BCR conformation during tumor evolution. We further followed evolutionary divergence by targeted sequencing of gene loci affected by aberrant SHM as well as of known driver genes of lymphomagenesis, and by array-based genome-wide chromosomal imbalance and DNA methylation analysis. We observed a wide spectrum of evolutionary patterns ranging from almost no evolution to divergent evolution within recurrent non-transformed t(14;18) FL. Remarkably, we observed a correlation of the magnitude of evolutionary divergence across all genetic and epigenetic levels suggesting co-evolution. The distribution of coding mutations in driver genes and the correlation with SHM suggest CREBBP and AID to be potential modifiers of genetic and epigenetic co-evolution in FL.