Polynomial algorithms for the Maximal Pairing Problem: Efficient phylogenetic targeting on arbitrary trees
 References:
References:
Arnold, C., Stadler, P.F. 2010. Polynomial algorithms for the Maximal Pairing Problem: Efficient phylogenetic targeting on arbitrary trees. Algorithms for Molecular Biology 5:25. [PDF]
Project Description:
For more information and an implementation of the program, see
http://www.bioinf.uni-leipzig.de/Software/Targeting/. In what follows, we provide an abstract of the project.
Background
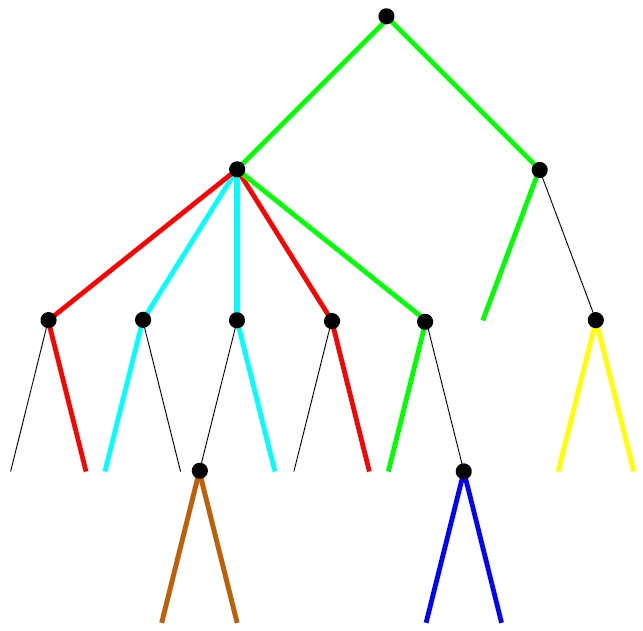
The Maximal Pairing Problem (MPP) is the prototype of a class of combinatorial optimization problems that are of considerable interest in bioinformatics: Given an arbitrary phylogenetic tree T and weights Ωxy for the paths between any two pairs of leaves (x, y), what is the collection of edge-disjoint paths between pairs of leaves that maximizes the total weight? Special cases of the MPP for binary trees and equal weights have been described previously. Algorithms to solve the general MPP are still missing, however.
Results
We describe a relatively simple dynamic programming algorithm for the special case of binary trees. The general case of multifurcating trees can then be treated by interleaving solutions to certain auxiliary Maximum Weighted Matching problems with an extension of this dynamic programming approach, resulting in an overall polynomial-time solution of complexity O(n4 log n) w.r.t. the number n of leaves. For binary trees, we furthermore discuss several constrained variants of the MPP as well as a partition function approach to the probabilistic version of the MPP.
Conclusions
The algorithms introduced here make it possible to solve the MPP also for large trees with high-degree vertices. This has practical relevance in the field of comparative phylogenetics and, for example, in the context of phylogenetic targeting, i.e., data collection with resource limitations.