Overview on processed synteny regions for SREBF2_ENSG00000198911_2e5.protein.info
| Rank | AlnOverview | AlnScore | RatioSum | logRatioSum | focalRatioSum | TargetGenome | chr | startPos | endPos | hitsCount | queryCov | meanIdentity | ori | score | compara |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 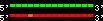 | 14000 | 1265 | 65.79 | 449.94 | Homo_sapiens | 22 | 40559221 | 40631607 | 18 | 94.7% | 95% | 1 | 2075.9 | within_species_paralog: SREBF2 |
| 2 | 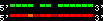 | 10327 | 161.03 | 25.76 | 35.93 | Macaca_mulatta | 10 | 85821766 | 85863757 | 15 | 84.4% | 91.4% | 1 | 1852.9 | ortholog_one2one: SREBF2 |
| 3 | 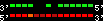 | 9002 | 82.71 | 18.94 | 24.42 | Canis_familiaris | 10 | 26516522 | 26577210 | 17 | 92.8% | 83.7% | -1 | 1836.7 | ortholog_one2one: 481228 |
| 4 | 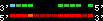 | 5522 | 47.71 | 8.89 | 14.19 | Gallus_gallus | 1 | 51370206 | 51385872 | 15 | 78.9% | 76.6% | -1 | 1531.3 | ortholog_one2one: Q9I8H7_CHICK |
| 5 |  | 1179 | 4.97 | 1.07 | 1.19 | Canis_familiaris | 10 | ||||||||
| 6 |  | 975 | 15 | 2.71 | 3.75 | Canis_familiaris | X | ||||||||
| 7 |  | 967 | 10 | 2.3 | 2.5 | Gasterosteus_aculeatus | contig_11123 | ||||||||
| 8 |  | 875 | 2.5 | 0.92 | 0.63 | Canis_familiaris | 24 | ||||||||
| 9 |  | 863 | 3.77 | 1.33 | 0.63 | Gasterosteus_aculeatus | contig_8218 | ||||||||
| 10 |  | 699 | 1 | 0 | 0.25 | Macaca_mulatta | 1 | ||||||||
| 11 |  | 699 | 1 | 0 | 0.25 | Homo_sapiens | 5 | ||||||||
| 12 |  | 665 | 0.91 | -0.08 | 0.23 | Canis_familiaris | X | ||||||||
| 13 |  | 661 | 1.79 | 0.58 | 0.22 | Gasterosteus_aculeatus | contig_8279 | ||||||||
| 14 |  | 606 | 1.51 | 0.41 | 0.19 | Gasterosteus_aculeatus | contig_9740 | ||||||||
| 15 |  | 519 | 1.02 | 0.02 | 0.17 | Gallus_gallus | 18 | ||||||||
| 16 |  | 495 | 2 | 0.69 | 1 | Gasterosteus_aculeatus | contig_4216 | ||||||||
| 17 |  | 481 | 0.59 | -0.52 | 0.15 | Macaca_mulatta | 11 | ||||||||
| 18 |  | 477 | 0.94 | -0.05 | 0.16 | Canis_familiaris | 6 | ||||||||
| 19 |  | 459 | 0.56 | -0.57 | 0.14 | Homo_sapiens | 12 | ||||||||
| 20 |  | 408 | 1 | 0 | 0.13 | Macaca_mulatta | X | ||||||||
| 21 |  | 368 | 0.94 | -0.05 | 0.12 | Gallus_gallus | 4 | ||||||||
| 22 |  | 338 | 0.74 | -0.29 | 0.12 | Homo_sapiens | 7 | ||||||||
| 23 |  | 333 | 1.17 | 0.16 | 1.17 | Gasterosteus_aculeatus | contig_4215 | 18120 | 24716 | 9 | 55% | 49.6% | -1 | 691.7 | noComparaHits: ENSGACG00000021431
; ENSGACG00000007259 |
| 24 |  | 331 | 1.09 | 0.09 | 0.12 | Gasterosteus_aculeatus | contig_2001 | ||||||||
| 25 |  | 314 | 1.13 | 0.12 | 1.13 | Gasterosteus_aculeatus | contig_4389 | 128552 | 135308 | 8 | 61.7% | 46.1% | 1 | 652.2 | noComparaHits: ENSGACG00000012064 |
| 26 |  | 312 | 0.72 | -0.32 | 0.12 | Homo_sapiens | 8 | ||||||||
| 27 |  | 307 | 0.72 | -0.32 | 0.12 | Gallus_gallus | 15 | ||||||||
| 28 |  | 301 | 1.04 | 0.04 | 0.12 | Gallus_gallus | 15 | ||||||||
| 29 |  | 298 | 0.7 | -0.35 | 0.12 | Gasterosteus_aculeatus | contig_5909 | ||||||||
| 30 |  | 297 | 0.7 | -0.35 | 0.12 | Homo_sapiens | 22 | ||||||||
| 31 |  | 293 | 0.7 | -0.35 | 0.12 | Macaca_mulatta | 15 | ||||||||
| 32 |  | 291 | 0.7 | -0.35 | 0.12 | Macaca_mulatta | 10 | ||||||||
| 33 |  | 277 | 0.68 | -0.38 | 0.11 | Gallus_gallus | 4 | ||||||||
| 34 |  | 275 | 1 | 0 | 0.11 | Macaca_mulatta | 11 | ||||||||
| 35 |  | 271 | 1 | 0 | 0.11 | Homo_sapiens | 12 | ||||||||
| 36 |  | 269 | 0.99 | 0 | 0.11 | Canis_familiaris | 26 | ||||||||
| 37 | 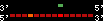 | 268 | 0.67 | -0.39 | 0.11 | Gallus_gallus | 1 | ||||||||
| 38 | 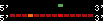 | 265 | 0.67 | -0.39 | 0.11 | Homo_sapiens | 9 | ||||||||
| 39 | 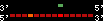 | 263 | 0.67 | -0.39 | 0.11 | Gallus_gallus | 13 | ||||||||
| 40 | 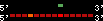 | 262 | 0.67 | -0.39 | 0.11 | Gallus_gallus | 4 | ||||||||
| 41 | 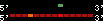 | 257 | 0.66 | -0.41 | 0.11 | Canis_familiaris | 32 | ||||||||
| 42 | 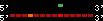 | 253 | 0.66 | -0.41 | 0.11 | Macaca_mulatta | 3 | ||||||||
| 43 | 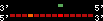 | 240 | 0.65 | -0.42 | 0.11 | Homo_sapiens | 16 | ||||||||
| 44 | 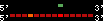 | 238 | 0.64 | -0.44 | 0.11 | Canis_familiaris | 6 | ||||||||
| 45 |  | 235 | 0.64 | -0.44 | 0.11 | Homo_sapiens | 19 | ||||||||
| 46 |  | 218 | 0.63 | -0.45 | 0.11 | Gasterosteus_aculeatus | contig_628 | ||||||||
| 47 |  | 216 | 0.99 | 0 | 0.99 | Macaca_mulatta | 16 | 17717122 | 17724076 | 6 | 40.8% | 50.9% | -1 | 448.8 | between_species_paralog: SREBF1 |
| 48 |  | 151 | 0.58 | -0.53 | 0.1 | Macaca_mulatta | 7 | ||||||||
| 49 |  | 149 | 0.58 | -0.53 | 0.1 | Canis_familiaris | 7 | ||||||||
| 50 |  | 97 | 0.86 | -0.14 | 0.86 | Gallus_gallus | 14 | 4933487 | 4934917 | 3 | 16.2% | 58.8% | -1 | 203.4 | noComparaHits: 416506 |
Data table for reference protein loci
QueryProtein(s)=ENSP00000354476 (contained in p4)QueryGenome=Homo_sapiens
| ref-loci | geneName | protID | transID | geneID | protName | chr | startPos | endPos | meanPos | ori | numExons | protLength | selfscore |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | NHP2L1 | ENSP00000347401 | ENST00000355257 | ENSG00000100138 | NH2L1_HUMAN | 22 | 40400883 | 40414746 | 40401072 | -1 | 3 | 128 | 228.6 |
| p2 | NP_689726.3 | ENSP00000300398 | ENST00000300398 | ENSG00000167077 | NP_689726.3 | 22 | 40425489 | 40525249 | 40479963 | 1 | 31 | 1280 | 2264.8 |
| p3 | CCDC134 | ENSP00000255784 | ENST00000255784 | ENSG00000100147 | CC134_HUMAN | 22 | 40534841 | 40551773 | 40539243 | 1 | 6 | 229 | 342.8 |
| p4 | SREBF2 | ENSP00000354476 | ENST00000361204 | ENSG00000198911 | SRBP2_HUMAN | 22 | 40559221 | 40631610 | 40604020 | 1 | 19 | 1141 | 2075.9 |
| Q8N8F0_HUMAN | ENSP00000327934 | ENST00000333487 | ENSG00000184068 | Q8N8F0_HUMAN | 22 | 40559573 | 40560424 | 40559762 | -1 | 2 | 129 | 140 | |
| p5 | Q5H8V2_HUMAN | ENSP00000370755 | ENST00000381351 | ENSG00000205705 | Q5H8V2_HUMAN | 22 | 40637198 | 40640516 | 40640247 | -1 | 2 | 177 | 129 |
| p6 | TNFRSF13C | ENSP00000291232 | ENST00000291232 | ENSG00000159958 | TR13C_HUMAN | 22 | 40651317 | 40652723 | 40652136 | -1 | 3 | 184 | 232.6 |
| p7 | CENPM | ENSP00000215980 | ENST00000215980 | ENSG00000100162 | CENPM_HUMAN | 22 | 40665006 | 40673026 | 40671209 | -1 | 6 | 180 | 317.7 |
| p8 | Q6ICG7_HUMAN | ENSP00000370752 | ENST00000381348 | ENSG00000205704 | Q6ICG7_HUMAN | 22 | 40684167 | 40684490 | 40684323 | 1 | 1 | 107 | 158 |
| p9 | SEPT3 | ENSP00000332866 | ENST00000328414 | ENSG00000100167 | SEPT3_HUMAN | 22 | 40707624 | 40722917 | 40715518 | 1 | 10 | 345 | 674.6 |
| p10 | WBP2NL | ENSP00000328800 | ENST00000329620 | ENSG00000183066 | NP_689826.1 | 22 | 40724769 | 40753131 | 40748252 | 1 | 6 | 309 | 323.1 |
| p11 | NAGA | ENSP00000355349 | ENST00000361920 | ENSG00000198951 | NAGAB_HUMAN | 22 | 40786229 | 40796247 | 40791823 | -1 | 9 | 411 | 893.5 |
| p12 | FAM109B | ENSP00000312753 | ENST00000321753 | ENSG00000177096 | F109B_HUMAN | 22 | 40803244 | 40804023 | 40803628 | 1 | 1 | 259 | 498 |
| p13 | C22orf32 | ENSP00000327467 | ENST00000331479 | ENSG00000183172 | NP_201575.2 | 22 | 40805719 | 40808012 | 40805875 | 1 | 2 | 107 | 186 |
| p14 | NDUFA6 | ENSP00000330937 | ENST00000331989 | ENSG00000184983 | NDUA6_HUMAN | 22 | 40812133 | 40816772 | 40813106 | -1 | 3 | 154 | 324.1 |
Parameter used for the loci-alignments
qLoci-Score_CutOff: 100 bitsqLoci-MaxProtOverlap: 40 aa
loci size factor: 2 (additional loci size constraint: 0 nt)
loci-grouping type: loci intervalls
max distance between non-highlighted (orange) target columns: 500000 nt
RelativeScore color index:
| <= 33 | <= 66 | <= 100 | <= 133 | <= 166 | <= 200 | <= 233 | <= 266 | <= 300 | <= 333 | <= 366 | <= 400 | <= 433 | <= 466 | <= 500 | <= 533 | <= 566 | <= 600 | <= 633 | <= 666 | <= 700 | <= 733 | <= 766 | <= 800 | <= 833 | <= 866 | <= 900 | <= 933 | <= 966 | <= 1000 | relSc>1000 |
region 1 (region 1 for Hsap)
 |
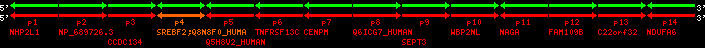 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 22 -i 40400886:40816772 -f 31e4 some_geneID_to_be_selected
scoreRatio_sum=1265 log_scoreRatio_sum=65.79 focal_scoreRatio_sum=449.94
AlnScore=14000 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 228.6 | 22 | 40400886 | 40406314 | -1 | 2 | 99.2% | 99.2% | 384 | 0.3 / 0.01 = 30 | within_species_paralog: NHP2L1 | 1 |
| p2 | q2 | +/+ | ENSP00000300398 | NP_689726.3 | q2 | q2 | 2264.8 | 22 | 40425489 | 40521615 | 1 | 25 | 93.3% | 91.3% | 3825 | 1 / 0.01 = 100 | noComparaHits: ENSG00000209534
; Y ; NP_689726.3 |
1 |
| p3 | q3 | +/+ | ENSP00000255784 | CCDC134 | q3 | q3 | 342.8 | 22 | 40535831 | 40539772 | 1 | 4 | 66.8% | 87.4% | 549 | 1 / 0.01 = 100 | noComparaHits: CCDC134 | 1 |
| p4 | q4 | ++/++ | ENSP00000354476 | SREBF2 | q4 | q4 | 2075.9 | 22 | 40559221 | 40631607 | 1 | 18 | 94.7% | 95% | 3402 | 0.77 / 0.01 = 77 | within_species_paralog: SREBF2 | 1 |
| ENSP00000327934 | Q8N8F0_HUMAN | q4 | 140 | 22 | 40559654 | 40559941 | -1 | 1 | 74.4% | 97.9% | 288 | 1 / 0.01 = 100 | noComparaHits: SREBF2
; Q8N8F0_HUMAN |
1 | ||||
| p5 | q5 | +/+ | ENSP00000370755 | Q5H8V2_HUMAN | q5 | q5 | 129 | 22 | 40640076 | 40640501 | -1 | 1 | 80.2% | 83.1% | 426 | 1 / 0.01 = 100 | noComparaHits: Q5H8V2_HUMAN | 1 |
| p6 | q6 | +/+ | ENSP00000291232 | TNFRSF13C | q6 | q6 | 232.6 | 22 | 40651320 | 40652723 | -1 | 3 | 97.8% | 82% | 633 | 1 / 0.01 = 100 | noComparaHits: TNFRSF13C | 1 |
| p7 | q7 | +/+ | ENSP00000215980 | CENPM | q7 | q7 | 317.7 | 22 | 40665009 | 40672446 | -1 | 5 | 89.4% | 90.7% | 546 | 1 / 0.01 = 100 | noComparaHits: CENPM | 1 |
| p8 | q8 | +/+ | ENSP00000370752 | Q6ICG7_HUMAN | q8 | q8 | 158 | 22 | 40684167 | 40684487 | 1 | 1 | 100% | 85% | 321 | 1 / 0.01 = 100 | noComparaHits: Q6ICG7_HUMAN | 1 |
| p9 | q9 | +/+ | ENSP00000332866 | SEPT3 | q9 | q9 | 674.6 | 22 | 40707624 | 40720664 | 1 | 8 | 91% | 88.3% | 1092 | 0.49 / 0.01 = 49 | within_species_paralog: SEPT3 | 1 |
| p10 | q10 | +/+ | ENSP00000328800 | WBP2NL | q10 | q10 | 323.1 | 22 | 40745259 | 40748317 | 1 | 4 | 50.2% | 92.4% | 510 | 0.78 / 0.01 = 78 | within_species_paralog: WBP2NL | 1 |
| p11 | q11 | +/+ | ENSP00000355349 | NAGA | q11 | q11 | 893.5 | 22 | 40786232 | 40794513 | -1 | 8 | 97.8% | 88.6% | 1401 | 0.59 / 0.01 = 59 | within_species_paralog: NAGA | 1 |
| p12 | q12 | +/+ | ENSP00000312753 | FAM109B | q12 | q12 | 498 | 22 | 40803244 | 40804020 | 1 | 1 | 100% | 100% | 777 | 0.72 / 0.01 = 72 | within_species_paralog: FAM109B | 1 |
| p13 | q13 | +/+ | ENSP00000327467 | C22orf32 | q13 | q13 | 186 | 22 | 40805737 | 40807991 | 1 | 2 | 88.8% | 97% | 300 | 1 / 0.01 = 100 | noComparaHits: C22orf32 | 1 |
| p14 | q14 | +/+ | ENSP00000330937 | NDUFA6 | q14 | q14 | 324.1 | 22 | 40812136 | 40816772 | -1 | 3 | 100% | 96.3% | 486 | 1 / 0.01 = 100 | noComparaHits: NDUFA6 | 1 |
region 2 (region 1 for Mmul)
 |
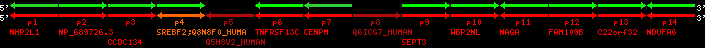 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 10 -i 85623310:86063311 -f 33e4 some_geneID_to_be_selected
scoreRatio_sum=161.03 log_scoreRatio_sum=25.76 focal_scoreRatio_sum=35.93
AlnScore=10327 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 147 | 10 | 85623310 | 85623570 | -1 | 1 | 68% | 100% | 261 | 0.3 / 0.35 = 0.86 | noComparaHits: | 2 |
| p2 | q2 | +/+ | ENSP00000300398 | NP_689726.3 | q2 | q2 | 1953.8 | 10 | 85647390 | 85749025 | 1 | 22 | 78.8% | 92.5% | 3213 | 1 / 0.13 = 7.69 | ortholog_one2one: NP_689726.3
ortholog_one2many: ENSMMUG00000031012 |
1 |
| p3 | q3 | +/+ | ENSP00000255784 | CCDC134 | q3 | q3 | 276.8 | 10 | 85762621 | 85766542 | 1 | 3 | 63.8% | 87.8% | 441 | 1 / 0.19 = 5.26 | ortholog_one2one: CCDC134 | 1 |
| p4 | q4 | +/+ | ENSP00000354476 | SREBF2 | q4 | q4 | 1852.9 | 10 | 85821766 | 85863757 | 1 | 15 | 84.4% | 91.4% | 3015 | 0.77 / 0.1 = 7.7 | ortholog_one2one: SREBF2 | 1 |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | noData | - | noData | ||||
| p5 | --- | gapq | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | q5 | +/+ | ENSP00000291232 | TNFRSF13C | q5 | q5 | 218.8 | 10 | 85883490 | 85884971 | -1 | 3 | 100% | 80.2% | 621 | 1 / 0.05 = 20 | ortholog_one2one: TNFRSF13C | 1 |
| p7 | q6 | +/+ | ENSP00000215980 | CENPM | q6 | q6 | 189.8 | 10 | 85903456 | 85906965 | -1 | 3 | 64.4% | 74.2% | 396 | 1 / 0.4 = 2.5 | ortholog_one2one: CENPM | 1 |
| p8 | --- | gapq | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q7 | +/+ | ENSP00000332866 | SEPT3 | q7 | q7 | 666.5 | 10 | 85944779 | 85958167 | 1 | 8 | 93.3% | 91.9% | 1026 | 0.49 / 0.01 = 49 | ortholog_one2one: SEPT3 | 1 |
| p10 | q8 | +/+ | ENSP00000328800 | WBP2NL | q8 | q8 | 297.4 | 10 | 85985108 | 85993649 | 1 | 4 | 50.2% | 85.3% | 510 | 0.78 / 0.07 = 11.14 | ortholog_one2one: WBP2NL | 1 |
| p11 | q9 | +/+ | ENSP00000355349 | NAGA | q9 | q9 | 785.7 | 10 | 86031454 | 86040392 | -1 | 7 | 89.1% | 84.5% | 1314 | 0.59 / 0.12 = 4.92 | ortholog_one2one: NAGA | 1 |
| p12 | q10 | +/+ | ENSP00000312753 | FAM109B | q10 | q10 | 459 | 10 | 86045748 | 86046524 | 1 | 1 | 100% | 93.8% | 777 | 0.72 / 0.07 = 10.29 | ortholog_one2one: FAM109B | 1 |
| p13 | q11 | +/+ | ENSP00000327467 | C22orf32 | q11 | q11 | 177.2 | 10 | 86048253 | 86050845 | 1 | 2 | 88.8% | 89.3% | 309 | 1 / 0.04 = 25 | ortholog_one2one: C22orf32 | 1 |
| p14 | q12 | +/+ | ENSP00000330937 | NDUFA6 | q12 | q12 | 302 | 10 | 86054698 | 86063311 | -1 | 3 | 100% | 93.1% | 477 | 1 / 0.06 = 16.67 | ortholog_one2one: NDUFA6 | 1 |
region 3 (region 1 for Cfam)
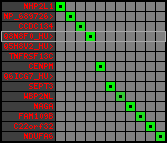 |
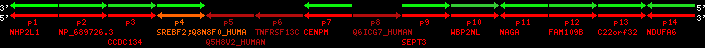 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 10 -i 26347775:26694189 -f 26e4 some_geneID_to_be_selected
scoreRatio_sum=82.71 log_scoreRatio_sum=18.94 focal_scoreRatio_sum=24.42
AlnScore=9002 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 227.5 | 10 | 26692968 | 26694189 | 1 | 2 | 99.2% | 97.7% | 390 | 0.3 / 0.01 = 30 | ortholog_one2one: 609886 | 1 |
| p2 | q2 | +/+ | ENSP00000300398 | NP_689726.3 | q2 | q2 | 1934.7 | 10 | 26598325 | 26670444 | -1 | 24 | 87.4% | 79.7% | 3690 | 1 / 0.14 = 7.14 | ortholog_one2one: NP_689726.3 | 1 |
| p3 | q3 | +/+ | ENSP00000255784 | CCDC134 | q3 | q3 | 260.3 | 10 | 26590778 | 26593513 | -1 | 3 | 56.3% | 87.1% | 420 | 1 / 0.24 = 4.17 | ortholog_one2one: 609694 | 1 |
| p4 | q4 | +/+ | ENSP00000354476 | SREBF2 | q4 | q4 | 1836.7 | 10 | 26516522 | 26577210 | -1 | 17 | 92.8% | 83.7% | 3384 | 0.77 / 0.11 = 7 | ortholog_one2one: 481228 | 1 |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | noData | - | noData | ||||
| p5 | --- | gapq | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gapq | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | q5 | +/+ | ENSP00000215980 | CENPM | q5 | q5 | 283.9 | 10 | 26477647 | 26488820 | 1 | 5 | 95.6% | 77.4% | 570 | 1 / 0.1 = 10 | ortholog_one2one: 609678 | 1 |
| p8 | --- | gapq | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q6 | +/+ | ENSP00000332866 | SEPT3 | q6 | q6 | 601.5 | 10 | 26437686 | 26449825 | -1 | 7 | 82.6% | 94.3% | 894 | 0.49 / 0.1 = 4.9 | ortholog_one2one: 481227 | 1 |
| p10 | q7 | +/+ | ENSP00000328800 | WBP2NL | q7 | q7 | 179.9 | 10 | 26417421 | 26418465 | -1 | 3 | 38.2% | 69.9% | 369 | 0.78 / 0.44 = 1.77 | ortholog_one2one: WBP2NL | 1 |
| p11 | q8 | +/+ | ENSP00000355349 | NAGA | q8 | q8 | 775.2 | 10 | 26369496 | 26375748 | 1 | 8 | 98.8% | 78.6% | 1317 | 0.59 / 0.13 = 4.54 | ortholog_one2one: 481226 | 1 |
| p12 | q9 | +/+ | ENSP00000312753 | FAM109B | q9 | q9 | 363 | 10 | 26362743 | 26363507 | -1 | 1 | 99.2% | 78.6% | 765 | 0.72 / 0.27 = 2.67 | noComparaHits: | 1 |
| p13 | q10 | +/+ | ENSP00000327467 | C22orf32 | q10 | q10 | 157.9 | 10 | 26358944 | 26360643 | -1 | 2 | 94.4% | 87.2% | 327 | 1 / 0.15 = 6.67 | ortholog_one2one: 609643 | 1 |
| p14 | q11 | +/+ | ENSP00000330937 | NDUFA6 | q11 | q11 | 237.7 | 10 | 26347775 | 26356193 | 1 | 3 | 100% | 78.8% | 447 | 1 / 0.26 = 3.85 | ortholog_one2one: 474483 | 1 |
region 4 (region 1 for Ggal)
 |
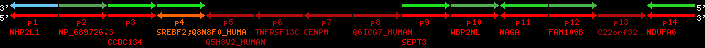 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 1 -i 51302418:51433031 -f 98e3 some_geneID_to_be_selected
scoreRatio_sum=47.71 log_scoreRatio_sum=8.89 focal_scoreRatio_sum=14.19
AlnScore=5522 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 232.1 | 1 | 51431916 | 51433031 | 1 | 2 | 99.2% | 88.8% | 402 | 0.3 / 0.01 = 30 | ortholog_one2one: NHP2L1 | 1 |
| p2 | q2 | +/+ | ENSP00000300398 | NP_689726.3 | q2 | q2 | 553.1 | 1 | 51396663 | 51425316 | -1 | 8 | 38.7% | 54.4% | 1581 | 1 / 0.75 = 1.33 | ortholog_one2one: NP_689726.3 | 1 |
| p3 | q3 | +/+ | ENSP00000255784 | CCDC134 | q3 | q3 | 261 | 1 | 51389704 | 51390703 | -1 | 3 | 71.6% | 57.9% | 657 | 1 / 0.23 = 4.35 | ortholog_one2one: 417984 | 1 |
| p4 | q4 | +/+ | ENSP00000354476 | SREBF2 | q4 | q4 | 1531.3 | 1 | 51370206 | 51385872 | -1 | 15 | 78.9% | 76.6% | 3069 | 0.77 / 0.26 = 2.96 | ortholog_one2one: Q9I8H7_CHICK | 1 |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | noData | - | noData | ||||
| p5 | --- | gapq | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gapq | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gapq | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gapq | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q5 | +/+ | ENSP00000332866 | SEPT3 | q5 | q5 | 502.8 | 1 | 51328295 | 51335085 | -1 | 6 | 71% | 90.6% | 768 | 0.49 / 0.25 = 1.96 | ortholog_one2one: 427899 | 1 |
| p10 | q6 | +/+ | ENSP00000328800 | WBP2NL | q6 | q6 | 105.1 | 1 | 51322448 | 51323546 | -1 | 2 | 30.1% | 55.1% | 288 | 0.78 / 0.67 = 1.16 | ortholog_one2one: Q5F3Y9_CHICK | 1 |
| p11 | q7 | +/+ | ENSP00000355349 | NAGA | q7 | q7 | 587.2 | 1 | 51312922 | 51315659 | 1 | 6 | 92.2% | 62.3% | 1335 | 0.59 / 0.34 = 1.74 | ortholog_one2one: NAGAB_CHICK | 1 |
| p12 | q8 | +/+ | ENSP00000312753 | FAM109B | q8 | q8 | 193 | 1 | 51310680 | 51311393 | -1 | 1 | 91.9% | 52.7% | 714 | 0.72 / 0.61 = 1.18 | ortholog_one2one: 427898 | 1 |
| p13 | --- | gapq | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | q9 | +/+ | ENSP00000330937 | NDUFA6 | q9 | q9 | 214.4 | 1 | 51302418 | 51303786 | 1 | 3 | 72.7% | 83.3% | 342 | 1 / 0.33 = 3.03 | ortholog_one2one: NDUFA6 | 1 |
region 5 (region 2 for Cfam)
 |
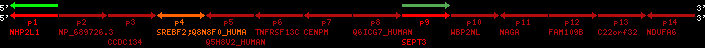 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 10 -i 37295114:37407905 -f 85e3 some_geneID_to_be_selected
scoreRatio_sum=4.97 log_scoreRatio_sum=1.07 focal_scoreRatio_sum=1.19
AlnScore=1179 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 211 | 10 | 37295114 | 37295473 | -1 | 1 | 93.8% | 94.2% | 360 | 0.3 / 0.07 = 4.29 | noComparaHits: | 3 |
| p2 | --- | gapq | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gapq | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gapq | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gapq | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gapq | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gapq | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gapq | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q2 | +/+ | ENSP00000332866 | SEPT3 | q2 | q2 | 182.2 | 10 | 37395972 | 37407905 | 1 | 3 | 53.6% | 46.2% | 543 | 0.49 / 0.72 = 0.68 | between_species_paralog: SEPT10 | 8 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
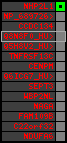
region 6 (region 3 for Cfam)
 |
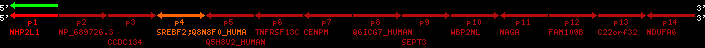 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l X -i 102573493:102573876 -f 29e1 some_geneID_to_be_selected
scoreRatio_sum=15 log_scoreRatio_sum=2.71 focal_scoreRatio_sum=3.75
AlnScore=975 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 223 | X | 102573493 | 102573876 | -1 | 1 | 100% | 98.4% | 384 | 0.3 / 0.02 = 15 | noComparaHits: ENSCAFG00000018660 | 2 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
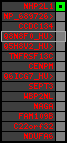
region 7 (region 1 for Gacu)
 |
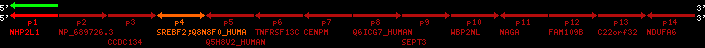 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_11123 -i 74912:75605 -f 52e1 some_geneID_to_be_selected
scoreRatio_sum=10 log_scoreRatio_sum=2.3 focal_scoreRatio_sum=2.5
AlnScore=967 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 221.2 | contig_11123 | 74912 | 75605 | -1 | 2 | 99.2% | 86.6% | 402 | 0.3 / 0.03 = 10 | noComparaHits: ENSGACG00000009568 | 1 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 8 (region 4 for Cfam)
 |
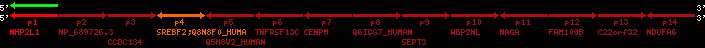 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 24 -i 35901840:35902261 -f 32e1 some_geneID_to_be_selected
scoreRatio_sum=2.5 log_scoreRatio_sum=0.92 focal_scoreRatio_sum=0.63
AlnScore=875 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 200.1 | 24 | 35901840 | 35902261 | -1 | 2 | 100% | 86% | 387 | 0.3 / 0.12 = 2.5 | ortholog_one2many: ENSCAFG00000009719 | 4 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 9 (region 2 for Gacu)
 |
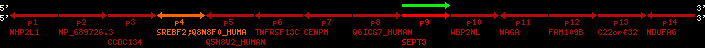 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_8218 -i 34510:81958 -f 36e3 some_geneID_to_be_selected
scoreRatio_sum=3.77 log_scoreRatio_sum=1.33 focal_scoreRatio_sum=0.63
AlnScore=863 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 582.6 | contig_8218 | 77831 | 81958 | 1 | 8 | 87.2% | 69% | 1182 | 0.49 / 0.13 = 3.77 | noComparaHits: ENSGACG00000015988 | 1 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 10 (region 2 for Mmul)
 |
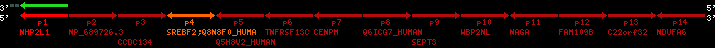 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 1 -i 127577156:128061284 -f 36e4 some_geneID_to_be_selected
scoreRatio_sum=1 log_scoreRatio_sum=0 focal_scoreRatio_sum=0.25
AlnScore=699 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| --- | q1 | gaps | - | - | - | q1 | - | - | - | - | - | - | - | - | - | - | - | - |
| --- | q2 | gaps | - | - | - | q2 | - | - | - | - | - | - | - | - | - | - | - | - |
| p1 | q3 | +/+ | ENSP00000347401 | NHP2L1 | q3 | q3 | 160 | 1 | 127577156 | 127577524 | 1 | 1 | 95.3% | 73% | 369 | 0.3 / 0.3 = 1 | noComparaHits: | 1 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 11 (region 2 for Hsap)
 |
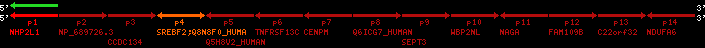 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 5 -i 96810875:96811201 -f 25e1 some_geneID_to_be_selected
scoreRatio_sum=1 log_scoreRatio_sum=0 focal_scoreRatio_sum=0.25
AlnScore=699 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 160 | 5 | 96810875 | 96811201 | -1 | 1 | 85.2% | 78.9% | 327 | 0.3 / 0.3 = 1 | noComparaHits: | 2 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 12 (region 5 for Cfam)
 |
 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l X -i 86848501:86848843 -f 26e1 some_geneID_to_be_selected
scoreRatio_sum=0.91 log_scoreRatio_sum=-0.08 focal_scoreRatio_sum=0.23
AlnScore=665 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 152.2 | X | 86848501 | 86848843 | -1 | 2 | 87.5% | 75.9% | 336 | 0.3 / 0.33 = 0.91 | noComparaHits: | 5 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 13 (region 3 for Gacu)
 |
 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_8279 -i 522:2619 -f 16e2 some_geneID_to_be_selected
scoreRatio_sum=1.79 log_scoreRatio_sum=0.58 focal_scoreRatio_sum=0.22
AlnScore=661 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | q1 | +/+ | ENSP00000355349 | NAGA | q1 | q1 | 591.2 | contig_8279 | 522 | 2619 | 1 | 7 | 94.4% | 59.4% | 1323 | 0.59 / 0.33 = 1.79 | noComparaHits: ENSGACG00000013389 | 1 |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 14 (region 4 for Gacu)
 |
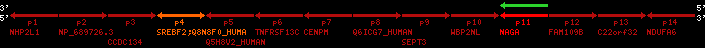 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_9740 -i 21958:27822 -f 44e2 some_geneID_to_be_selected
scoreRatio_sum=1.51 log_scoreRatio_sum=0.41 focal_scoreRatio_sum=0.19
AlnScore=606 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | q1 | +/+ | ENSP00000355349 | NAGA | q1 | q1 | 542.1 | contig_9740 | 21958 | 27822 | 1 | 7 | 96.4% | 53.2% | 1404 | 0.59 / 0.39 = 1.51 | noComparaHits: ENSGACG00000014083 | 2 |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 15 (region 2 for Ggal)
 |
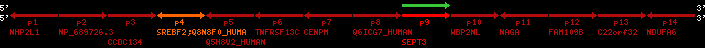 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 18 -i 3800336:3808951 -f 65e2 some_geneID_to_be_selected
scoreRatio_sum=1.02 log_scoreRatio_sum=0.02 focal_scoreRatio_sum=0.17
AlnScore=519 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 350.4 | 18 | 3800336 | 3808951 | 1 | 5 | 62.9% | 70.3% | 654 | 0.49 / 0.48 = 1.02 | between_species_paralog: Q5F3T2_CHICK | 2 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 16 (region 5 for Gacu)
 |
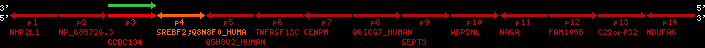 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_4216 -i 4884:6155 -f 95e1 some_geneID_to_be_selected
scoreRatio_sum=2 log_scoreRatio_sum=0.69 focal_scoreRatio_sum=1
AlnScore=495 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | q1 | +/+ | ENSP00000255784 | CCDC134 | q1 | q1 | 169.9 | contig_4216 | 4884 | 6155 | -1 | 2 | 44.5% | 69.6% | 306 | 1 / 0.5 = 2 | noComparaHits: ENSGACG00000007268 | 1 |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 17 (region 3 for Mmul)
 |
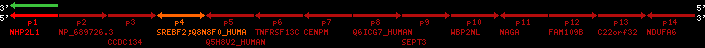 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 11 -i 134176583:134176965 -f 29e1 some_geneID_to_be_selected
scoreRatio_sum=0.59 log_scoreRatio_sum=-0.52 focal_scoreRatio_sum=0.15
AlnScore=481 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 110 | 11 | 134176583 | 134176900 | 1 | 1 | 80.5% | 61.7% | 318 | 0.3 / 0.51 = 0.59 | noComparaHits: | 5 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 18 (region 6 for Cfam)
 |
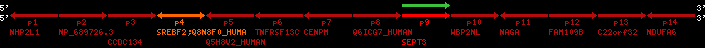 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 6 -i 39676207:39683553 -f 55e2 some_geneID_to_be_selected
scoreRatio_sum=0.94 log_scoreRatio_sum=-0.05 focal_scoreRatio_sum=0.16
AlnScore=477 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 321.8 | 6 | 39676207 | 39683553 | 1 | 5 | 73.9% | 48.9% | 939 | 0.49 / 0.52 = 0.94 | between_species_paralog: NP_653206.1 | 3 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 19 (region 3 for Hsap)
 |
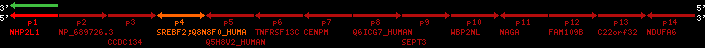 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 12 -i 131986645:131987027 -f 29e1 some_geneID_to_be_selected
scoreRatio_sum=0.56 log_scoreRatio_sum=-0.57 focal_scoreRatio_sum=0.14
AlnScore=459 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | q1 | +/+ | ENSP00000347401 | NHP2L1 | q1 | q1 | 105 | 12 | 131986645 | 131986962 | 1 | 1 | 80.5% | 60.7% | 318 | 0.3 / 0.54 = 0.56 | noComparaHits: | 3 |
| p2 | --- | gape | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gape | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gape | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 20 (region 4 for Mmul)
 |
 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l X -i 100116244:100125581 -f 70e2 some_geneID_to_be_selected
scoreRatio_sum=1 log_scoreRatio_sum=0 focal_scoreRatio_sum=0.13
AlnScore=408 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | q1 | +/+ | ENSP00000355349 | NAGA | q1 | q1 | 364.8 | X | 100116244 | 100125581 | -1 | 5 | 74.9% | 53% | 1011 | 0.59 / 0.59 = 1 | between_species_paralog: GLA | 2 |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 21 (region 3 for Ggal)
 |
 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 4 -i 1999508:2005441 -f 45e2 some_geneID_to_be_selected
scoreRatio_sum=0.94 log_scoreRatio_sum=-0.05 focal_scoreRatio_sum=0.12
AlnScore=368 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | q1 | +/+ | ENSP00000355349 | NAGA | q1 | q1 | 329.4 | 4 | 1999508 | 2005441 | 1 | 4 | 63% | 52.6% | 864 | 0.59 / 0.63 = 0.94 | between_species_paralog: 422188 | 2 |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 22 (region 4 for Hsap)
 |
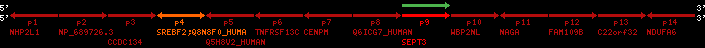 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 7 -i 151695662:151696310 -f 49e1 some_geneID_to_be_selected
scoreRatio_sum=0.74 log_scoreRatio_sum=-0.29 focal_scoreRatio_sum=0.12
AlnScore=338 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 228.6 | 7 | 151695662 | 151696310 | 1 | 2 | 63.5% | 49.3% | 648 | 0.49 / 0.66 = 0.74 | noComparaHits: MLL3 | 4 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 23 (region 6 for Gacu)
 |
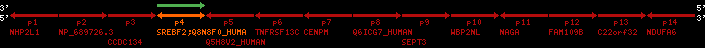 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_4215 -i 18120:24716 -f 49e2 some_geneID_to_be_selected
scoreRatio_sum=1.17 log_scoreRatio_sum=0.16 focal_scoreRatio_sum=1.17
AlnScore=333 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | q1 | +/+ | ENSP00000354476 | SREBF2 | q1 | q1 | 691.7 | contig_4215 | 18120 | 24716 | -1 | 9 | 55% | 49.6% | 2229 | 0.77 / 0.66 = 1.17 | noComparaHits: ENSGACG00000021431
; ENSGACG00000007259 |
1 |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | noData | - | noData | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 24 (region 7 for Gacu)
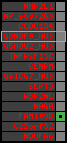 |
 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_2001 -i 83219:83995 -f 58e1 some_geneID_to_be_selected
scoreRatio_sum=1.09 log_scoreRatio_sum=0.09 focal_scoreRatio_sum=0.12
AlnScore=331 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gaps | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | q1 | +/+ | ENSP00000312753 | FAM109B | q1 | q1 | 165 | contig_2001 | 83219 | 83995 | -1 | 1 | 93.4% | 43.8% | 777 | 0.72 / 0.66 = 1.09 | noComparaHits: ENSGACG00000006284 | 1 |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 25 (region 8 for Gacu)
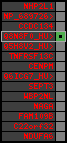 |
 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_4389 -i 128552:135308 -f 51e2 some_geneID_to_be_selected
scoreRatio_sum=1.13 log_scoreRatio_sum=0.12 focal_scoreRatio_sum=1.13
AlnScore=314 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | q1 | +/+ | ENSP00000354476 | SREBF2 | q1 | q1 | 652.2 | contig_4389 | 128552 | 135308 | 1 | 8 | 61.7% | 46.1% | 2268 | 0.77 / 0.68 = 1.13 | noComparaHits: ENSGACG00000012064 | 2 |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | noData | - | noData | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 26 (region 5 for Hsap)
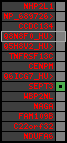 |
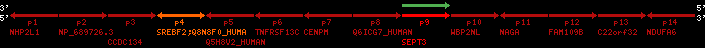 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 8 -i 57552312:57553100 -f 59e1 some_geneID_to_be_selected
scoreRatio_sum=0.72 log_scoreRatio_sum=-0.32 focal_scoreRatio_sum=0.12
AlnScore=312 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 211 | 8 | 57552312 | 57553100 | -1 | 1 | 78.3% | 43.5% | 789 | 0.49 / 0.68 = 0.72 | noComparaHits: | 5 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 27 (region 4 for Ggal)
 |
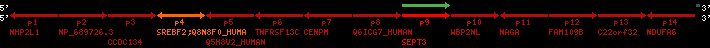 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 15 -i 724373:782404 -f 44e3 some_geneID_to_be_selected
scoreRatio_sum=0.72 log_scoreRatio_sum=-0.32 focal_scoreRatio_sum=0.12
AlnScore=307 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 214.5 | 15 | 724373 | 733262 | 1 | 4 | 50.7% | 53.3% | 543 | 0.49 / 0.68 = 0.72 | between_species_paralog: Q5ZMH1_CHICK | 3 |
| p10 | --- | gapq | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gapq | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gapq | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gapq | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gapq | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| --- | q2 | gape | - | - | - | q2 | - | - | - | - | - | - | - | - | - | - | - | - |
region 28 (region 5 for Ggal)
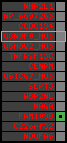 |
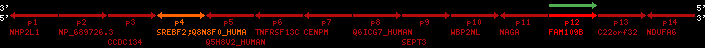 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 15 -i 6149980:6150675 -f 52e1 some_geneID_to_be_selected
scoreRatio_sum=1.04 log_scoreRatio_sum=0.04 focal_scoreRatio_sum=0.12
AlnScore=301 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gaps | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | q1 | +/+ | ENSP00000312753 | FAM109B | q1 | q1 | 150 | 15 | 6149980 | 6150675 | -1 | 1 | 94.6% | 42.6% | 696 | 0.72 / 0.69 = 1.04 | between_species_paralog: 427697 | 2 |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 29 (region 9 for Gacu)
 |
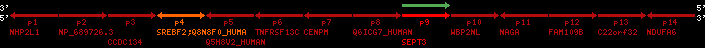 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_5909 -i 36857:38584 -f 13e2 some_geneID_to_be_selected
scoreRatio_sum=0.7 log_scoreRatio_sum=-0.35 focal_scoreRatio_sum=0.12
AlnScore=298 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 201.1 | contig_5909 | 36857 | 38584 | -1 | 3 | 73.6% | 35.3% | 1047 | 0.49 / 0.7 = 0.7 | noComparaHits: ENSGACG00000018181 | 10 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 30 (region 6 for Hsap)
 |
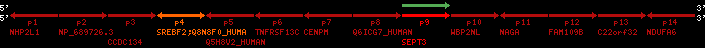 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 22 -i 18087637:18160392 -f 55e3 some_geneID_to_be_selected
scoreRatio_sum=0.7 log_scoreRatio_sum=-0.35 focal_scoreRatio_sum=0.12
AlnScore=297 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 201 | 22 | 18087637 | 18089457 | 1 | 3 | 53.3% | 47.7% | 639 | 0.49 / 0.7 = 0.7 | within_species_paralog: SEPT5 | 7 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 31 (region 5 for Mmul)
 |
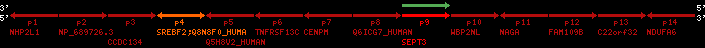 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 15 -i 37624124:37624895 -f 58e1 some_geneID_to_be_selected
scoreRatio_sum=0.7 log_scoreRatio_sum=-0.35 focal_scoreRatio_sum=0.12
AlnScore=293 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 198.2 | 15 | 37624124 | 37624895 | -1 | 2 | 80.6% | 38.4% | 717 | 0.49 / 0.7 = 0.7 | between_species_paralog: ENSMMUG00000006807 | 6 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 32 (region 6 for Mmul)
 |
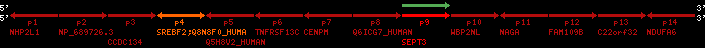 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 10 -i 63768564:63770385 -f 14e2 some_geneID_to_be_selected
scoreRatio_sum=0.7 log_scoreRatio_sum=-0.35 focal_scoreRatio_sum=0.12
AlnScore=291 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 196.8 | 10 | 63768564 | 63770385 | 1 | 3 | 53.3% | 46.7% | 639 | 0.49 / 0.7 = 0.7 | between_species_paralog: SEPT5 | 7 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 33 (region 6 for Ggal)
 |
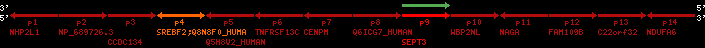 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 4 -i 51161077:51172153 -f 83e2 some_geneID_to_be_selected
scoreRatio_sum=0.68 log_scoreRatio_sum=-0.38 focal_scoreRatio_sum=0.11
AlnScore=277 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 187.2 | 4 | 51161077 | 51172153 | -1 | 3 | 54.2% | 46.1% | 558 | 0.49 / 0.72 = 0.68 | between_species_paralog: SEPT11 | 6 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 34 (region 7 for Mmul)
 |
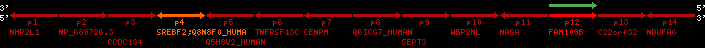 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 11 -i 112301009:112301401 -f 29e1 some_geneID_to_be_selected
scoreRatio_sum=1 log_scoreRatio_sum=0 focal_scoreRatio_sum=0.11
AlnScore=275 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gaps | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | q1 | +/+ | ENSP00000312753 | FAM109B | q1 | q1 | 137 | 11 | 112301009 | 112301401 | -1 | 1 | 53.7% | 55.4% | 393 | 0.72 / 0.72 = 1 | between_species_paralog: FAM109A | 2 |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 35 (region 7 for Hsap)
 |
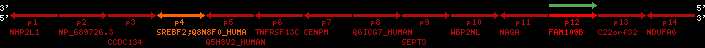 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 12 -i 110285222:110285614 -f 29e1 some_geneID_to_be_selected
scoreRatio_sum=1 log_scoreRatio_sum=0 focal_scoreRatio_sum=0.11
AlnScore=271 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gaps | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | q1 | +/+ | ENSP00000312753 | FAM109B | q1 | q1 | 135 | 12 | 110285222 | 110285614 | -1 | 1 | 53.7% | 54.7% | 393 | 0.72 / 0.72 = 1 | within_species_paralog: FAM109A | 2 |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 36 (region 7 for Cfam)
 |
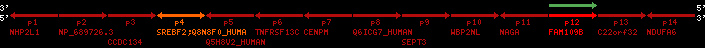 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 26 -i 12023153:12023533 -f 29e1 some_geneID_to_be_selected
scoreRatio_sum=0.99 log_scoreRatio_sum=0 focal_scoreRatio_sum=0.11
AlnScore=269 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gaps | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gaps | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gaps | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | q1 | +/+ | ENSP00000312753 | FAM109B | q1 | q1 | 134 | 26 | 12023153 | 12023533 | -1 | 1 | 52.1% | 57% | 381 | 0.72 / 0.73 = 0.99 | ortholog_one2many: 486268 | 2 |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 37 (region 7 for Ggal)
 |
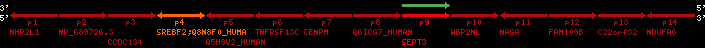 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 1 -i 141061311:141069115 -f 59e2 some_geneID_to_be_selected
scoreRatio_sum=0.67 log_scoreRatio_sum=-0.39 focal_scoreRatio_sum=0.11
AlnScore=268 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 181.4 | 1 | 141061311 | 141069115 | -1 | 3 | 52.8% | 47% | 534 | 0.49 / 0.73 = 0.67 | between_species_paralog: 418732 | 7 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 38 (region 8 for Hsap)
 |
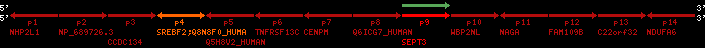 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 9 -i 100409227:100409867 -f 48e1 some_geneID_to_be_selected
scoreRatio_sum=0.67 log_scoreRatio_sum=-0.39 focal_scoreRatio_sum=0.11
AlnScore=265 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 179.2 | 9 | 100409227 | 100409867 | 1 | 2 | 55.1% | 44.1% | 558 | 0.49 / 0.73 = 0.67 | noComparaHits: GABBR2 | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 39 (region 8 for Ggal)
 |
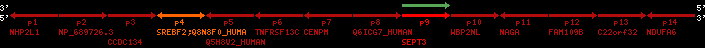 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 13 -i 17576574:17585447 -f 67e2 some_geneID_to_be_selected
scoreRatio_sum=0.67 log_scoreRatio_sum=-0.39 focal_scoreRatio_sum=0.11
AlnScore=263 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 177.9 | 13 | 17576574 | 17585447 | -1 | 3 | 52.5% | 45.1% | 531 | 0.49 / 0.73 = 0.67 | between_species_paralog: 416333 | 8 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 40 (region 9 for Ggal)
 |
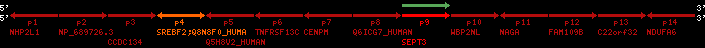 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 4 -i 16648292:16655179 -f 52e2 some_geneID_to_be_selected
scoreRatio_sum=0.67 log_scoreRatio_sum=-0.39 focal_scoreRatio_sum=0.11
AlnScore=262 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 176.8 | 4 | 16648292 | 16655179 | 1 | 3 | 54.8% | 45.5% | 558 | 0.49 / 0.73 = 0.67 | between_species_paralog: Q5ZM42_CHICK | 9 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 41 (region 8 for Cfam)
 |
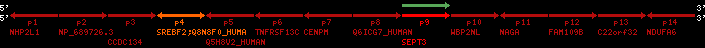 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 32 -i 27989317:27990155 -f 63e1 some_geneID_to_be_selected
scoreRatio_sum=0.66 log_scoreRatio_sum=-0.41 focal_scoreRatio_sum=0.11
AlnScore=257 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 174 | 32 | 27989317 | 27989844 | 1 | 1 | 51.3% | 48.6% | 528 | 0.49 / 0.74 = 0.66 | noComparaHits: ENSCAFG00000010861 | 10 |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 42 (region 8 for Mmul)
 |
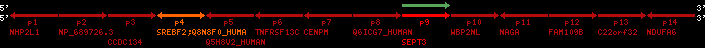 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 3 -i 189312557:189313190 -f 48e1 some_geneID_to_be_selected
scoreRatio_sum=0.66 log_scoreRatio_sum=-0.41 focal_scoreRatio_sum=0.11
AlnScore=253 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 171 | 3 | 189312696 | 189313190 | 1 | 1 | 48.4% | 50.3% | 495 | 0.49 / 0.74 = 0.66 | between_species_paralog: ENSMMUG00000007222 | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 43 (region 9 for Hsap)
 |
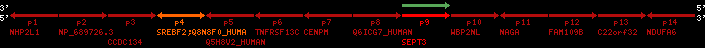 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 16 -i 30275249:30300294 -f 19e3 some_geneID_to_be_selected
scoreRatio_sum=0.65 log_scoreRatio_sum=-0.42 focal_scoreRatio_sum=0.11
AlnScore=240 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 162.5 | 16 | 30297884 | 30300294 | -1 | 3 | 36.5% | 55.6% | 375 | 0.49 / 0.75 = 0.65 | within_species_paralog: SEPT1 | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 44 (region 9 for Cfam)
 |
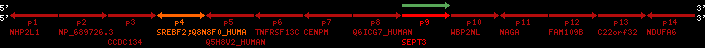 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 6 -i 20729320:20748791 -f 15e3 some_geneID_to_be_selected
scoreRatio_sum=0.64 log_scoreRatio_sum=-0.44 focal_scoreRatio_sum=0.11
AlnScore=238 (based on relative scores)
AlnType: normal (increasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 161 | 6 | 20729320 | 20731187 | 1 | 3 | 36.8% | 54.3% | 384 | 0.49 / 0.76 = 0.64 | between_species_paralog: 489900 | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 45 (region 10 for Hsap)
 |
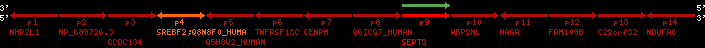 |
retrieve target region: getEnsemblProteins.pl -h Homo_sapiens -l 19 -i 58967767:58968361 -f 45e1 some_geneID_to_be_selected
scoreRatio_sum=0.64 log_scoreRatio_sum=-0.44 focal_scoreRatio_sum=0.11
AlnScore=235 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Homo_sapiens
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 159.1 | 19 | 58967767 | 58968361 | -1 | 2 | 56.2% | 41.1% | 585 | 0.49 / 0.76 = 0.64 | noComparaHits: | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 46 (region 10 for Gacu)
 |
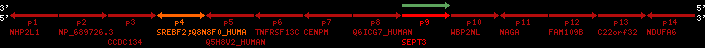 |
retrieve target region: getEnsemblProteins.pl -h Gasterosteus_aculeatus -l contig_628 -i 122373:123379 -f 76e1 some_geneID_to_be_selected
scoreRatio_sum=0.63 log_scoreRatio_sum=-0.45 focal_scoreRatio_sum=0.11
AlnScore=218 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gasterosteus_aculeatus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 147.5 | contig_628 | 122373 | 123379 | -1 | 2 | 36.2% | 45.5% | 459 | 0.49 / 0.78 = 0.63 | noComparaHits: ENSGACG00000020488 | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 47 (region 9 for Mmul)
 |
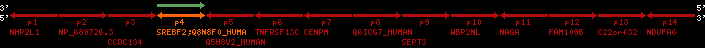 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 16 -i 17717122:17724076 -f 52e2 some_geneID_to_be_selected
scoreRatio_sum=0.99 log_scoreRatio_sum=0 focal_scoreRatio_sum=0.99
AlnScore=216 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | q1 | +/+ | ENSP00000354476 | SREBF2 | q1 | q1 | 448.8 | 16 | 17717122 | 17724076 | -1 | 6 | 40.8% | 50.9% | 1506 | 0.77 / 0.78 = 0.99 | between_species_paralog: SREBF1 | 2 |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | noData | - | noData | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 48 (region 10 for Mmul)
 |
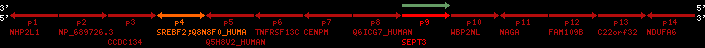 |
retrieve target region: getEnsemblProteins.pl -h Macaca_mulatta -l 7 -i 98116277:98116708 -f 32e1 some_geneID_to_be_selected
scoreRatio_sum=0.58 log_scoreRatio_sum=-0.53 focal_scoreRatio_sum=0.1
AlnScore=151 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Macaca_mulatta
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 102 | 7 | 98116277 | 98116708 | -1 | 1 | 42.3% | 36.7% | 432 | 0.49 / 0.84 = 0.58 | noComparaHits: KIAA0391 | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 49 (region 10 for Cfam)
 |
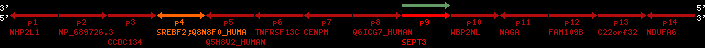 |
retrieve target region: getEnsemblProteins.pl -h Canis_familiaris -l 7 -i 81393240:81393593 -f 27e1 some_geneID_to_be_selected
scoreRatio_sum=0.58 log_scoreRatio_sum=-0.53 focal_scoreRatio_sum=0.1
AlnScore=149 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Canis_familiaris
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | --- | gaps | ENSP00000354476 | SREBF2 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | - | - | - | - | - | - | - | - | - | - | - | - | ||||
| p5 | --- | gaps | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gaps | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gaps | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gaps | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | q1 | +/+ | ENSP00000332866 | SEPT3 | q1 | q1 | 101 | 7 | 81393240 | 81393593 | -1 | 1 | 35.9% | 41.9% | 354 | 0.49 / 0.85 = 0.58 | noComparaHits: | noData |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
region 50 (region 10 for Ggal)
 |
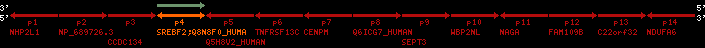 |
retrieve target region: getEnsemblProteins.pl -h Gallus_gallus -l 14 -i 4933487:4934917 -f 11e2 some_geneID_to_be_selected
scoreRatio_sum=0.86 log_scoreRatio_sum=-0.14 focal_scoreRatio_sum=0.86
AlnScore=97 (based on relative scores)
AlnType: reversed (decreasing chromosome positions)
QueryProtein(s)=ENSP00000354476 (contained in p4)
TargetGenome=Gallus_gallus
| ref-loci | target-loci | aln | protID | geneName | optQloci | target-loci | score | chr | startPos | endPos | ori | hits | queryCov | meanIdentity | hitsLength | intra_inter | compara | globalRank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p1 | --- | gaps | ENSP00000347401 | NHP2L1 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p2 | --- | gaps | ENSP00000300398 | NP_689726.3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p3 | --- | gaps | ENSP00000255784 | CCDC134 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p4 | q1 | +/+ | ENSP00000354476 | SREBF2 | q1 | q1 | 203.4 | 14 | 4933487 | 4934917 | -1 | 3 | 16.2% | 58.8% | 558 | 0.77 / 0.9 = 0.86 | noComparaHits: 416506 | 2 |
| ENSP00000327934 | Q8N8F0_HUMAN | noMatch | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | NoData | noData | - | noData | ||||
| p5 | --- | gape | ENSP00000370755 | Q5H8V2_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p6 | --- | gape | ENSP00000291232 | TNFRSF13C | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p7 | --- | gape | ENSP00000215980 | CENPM | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p8 | --- | gape | ENSP00000370752 | Q6ICG7_HUMAN | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p9 | --- | gape | ENSP00000332866 | SEPT3 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p10 | --- | gape | ENSP00000328800 | WBP2NL | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p11 | --- | gape | ENSP00000355349 | NAGA | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p12 | --- | gape | ENSP00000312753 | FAM109B | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p13 | --- | gape | ENSP00000327467 | C22orf32 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |
| p14 | --- | gape | ENSP00000330937 | NDUFA6 | noMatch | --- | - | - | - | - | - | - | - | - | - | - | - | - |