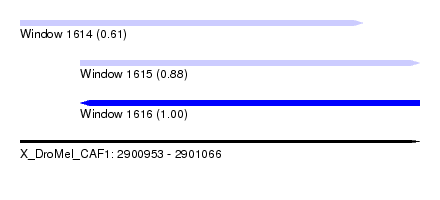
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,900,953 – 2,901,066 |
| Length | 113 |
| Max. P | 0.995693 |

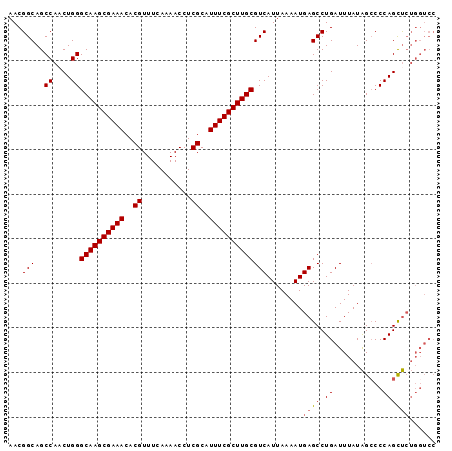
| Location | 2,900,953 – 2,901,050 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2900953 97 + 22224390 -----------------------GGAGAAUCAAAUUGCAAAACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUA -----------------------...((((((..((((......))))......((((((((((((..((...........))..)))))))))).((((....)))).))))))))... ( -24.60) >DroPse_CAF1 394691 97 + 1 -----------------------AGAGAAUCAAAUUGCAAAACAGCAGCCAACUGGGAAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAAGAGCCUGAUUUAUA -----------------------...((((((..((((......))))((....))..((((((((..((...........))..))))))))((.((........)))).))))))... ( -16.80) >DroEre_CAF1 311390 97 + 1 -----------------------GGAGAAUCAAAUUGCAAAACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUA -----------------------...((((((..((((......))))......((((((((((((..((...........))..)))))))))).((((....)))).))))))))... ( -24.60) >DroYak_CAF1 295999 97 + 1 -----------------------GGAGAAUCAAAUUGCAAAACGGCAGCCAUCUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUA -----------------------...((((((..((((......))))......((((((((((((..((...........))..)))))))))).((((....)))).))))))))... ( -24.60) >DroAna_CAF1 454231 120 + 1 GGGGGGCACCGCCACCCCACCGAAGAGAAUCAAAUUGCAAAACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUG (((((((...))).))))........((((((..((((......))))......((((((((((((..((...........))..)))))))))).((((....)))).))))))))... ( -36.70) >DroPer_CAF1 415192 97 + 1 -----------------------AGAGAAUCAAAUUGCAAAACAGCAGCCAACUGGGAAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAAGAGCCUGAUUUAUA -----------------------...((((((..((((......))))((....))..((((((((..((...........))..))))))))((.((........)))).))))))... ( -16.80) >consensus _______________________AGAGAAUCAAAUUGCAAAACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUA ..........................((((((..((((......))))......((((((((((((..((...........))..)))))))))).((((....)))).))))))))... (-21.07 = -21.73 + 0.67)

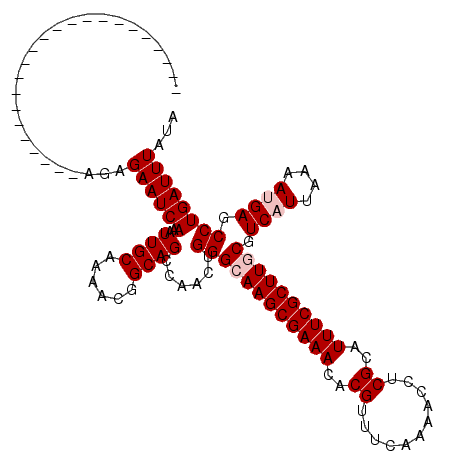
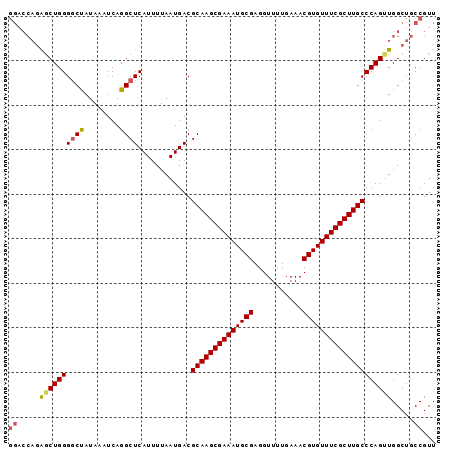
| Location | 2,900,970 – 2,901,066 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2900970 96 + 22224390 AACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUAGCCCCAGCUCUGGUCC ...((..((((.((((((((((((((..((...........))..)))))))))).((((....))))...............))))...)))))) ( -26.00) >DroSec_CAF1 287755 96 + 1 AACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUAGCCCCAGCUCUGGUCC ...((..((((.((((((((((((((..((...........))..)))))))))).((((....))))...............))))...)))))) ( -26.00) >DroSim_CAF1 258046 96 + 1 AACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUAGCCCCAGCUCUGGUCC ...((..((((.((((((((((((((..((...........))..)))))))))).((((....))))...............))))...)))))) ( -26.00) >DroEre_CAF1 311407 96 + 1 AACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUAGACCCAGCUCUGGUCC ...(((..((....))((((((((((..((...........))..)))))))))).((((....))))))).........((((.......)))). ( -26.60) >DroYak_CAF1 296016 96 + 1 AACGGCAGCCAUCUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUAGCCCCAGCUCUGUUCC (((((.(((.....((((((((((((..((...........))..))))))))((.((((....))))))..........))))..)))))))).. ( -25.90) >DroAna_CAF1 454271 96 + 1 AACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUGGCCCCAGUCAGUGGGA ........((((((((((((((((((..((...........))..)))))))))).((((....))))(((........))).)))))...))).. ( -29.80) >consensus AACGGCAGCCAACUGGGCAAGCGAAACACGUUUCAAAACCUCGCAUUUCGCUUGCGUCAUUAAAAUGAGCCUGAUUUAUAGCCCCAGCUCUGGUCC ...(((..((....))((((((((((..((...........))..)))))))))))))........((((.((...........))))))...... (-23.32 = -23.52 + 0.19)

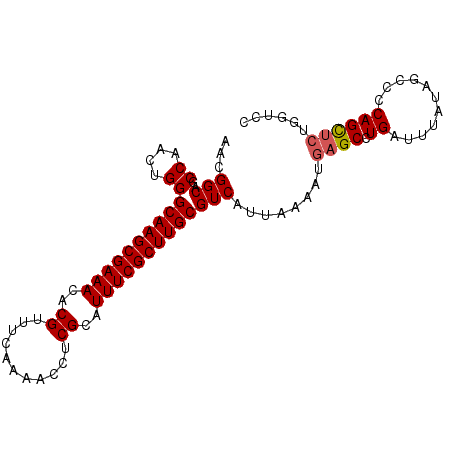
| Location | 2,900,970 – 2,901,066 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -30.57 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
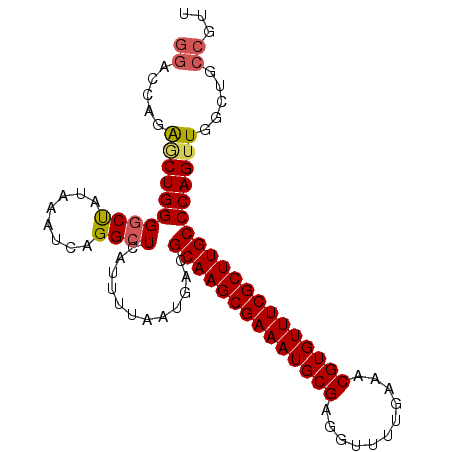
>X_DroMel_CAF1 2900970 96 - 22224390 GGACCAGAGCUGGGGCUAUAAAUCAGGCUCAUUUUAAUGACGCAAGCGAAAUGCGAGGUUUUGAAACGUGUUUCGCUUGCCCAGUUGGCUGCCGUU ((.(((..(((((((((........))))............((((((((((((((...........))))))))))))))))))))))...))... ( -34.60) >DroSec_CAF1 287755 96 - 1 GGACCAGAGCUGGGGCUAUAAAUCAGGCUCAUUUUAAUGACGCAAGCGAAAUGCGAGGUUUUGAAACGUGUUUCGCUUGCCCAGUUGGCUGCCGUU ((.(((..(((((((((........))))............((((((((((((((...........))))))))))))))))))))))...))... ( -34.60) >DroSim_CAF1 258046 96 - 1 GGACCAGAGCUGGGGCUAUAAAUCAGGCUCAUUUUAAUGACGCAAGCGAAAUGCGAGGUUUUGAAACGUGUUUCGCUUGCCCAGUUGGCUGCCGUU ((.(((..(((((((((........))))............((((((((((((((...........))))))))))))))))))))))...))... ( -34.60) >DroEre_CAF1 311407 96 - 1 GGACCAGAGCUGGGUCUAUAAAUCAGGCUCAUUUUAAUGACGCAAGCGAAAUGCGAGGUUUUGAAACGUGUUUCGCUUGCCCAGUUGGCUGCCGUU ((.(((..((((((((..((((..........))))..)))((((((((((((((...........))))))))))))))))))))))...))... ( -33.90) >DroYak_CAF1 296016 96 - 1 GGAACAGAGCUGGGGCUAUAAAUCAGGCUCAUUUUAAUGACGCAAGCGAAAUGCGAGGUUUUGAAACGUGUUUCGCUUGCCCAGAUGGCUGCCGUU ((..(((..((((((((........))))............((((((((((((((...........))))))))))))))))))....)))))... ( -32.70) >DroAna_CAF1 454271 96 - 1 UCCCACUGACUGGGGCCAUAAAUCAGGCUCAUUUUAAUGACGCAAGCGAAAUGCGAGGUUUUGAAACGUGUUUCGCUUGCCCAGUUGGCUGCCGUU ...(((..(((((((((........))))............((((((((((((((...........)))))))))))))))))))..).))..... ( -36.20) >consensus GGACCAGAGCUGGGGCUAUAAAUCAGGCUCAUUUUAAUGACGCAAGCGAAAUGCGAGGUUUUGAAACGUGUUUCGCUUGCCCAGUUGGCUGCCGUU ((.....((((((((((........))))............((((((((((((((...........)))))))))))))))))))).....))... (-30.57 = -30.85 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:00 2006