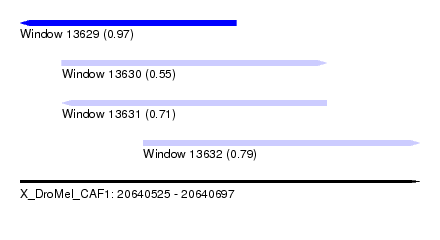
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,640,525 – 20,640,697 |
| Length | 172 |
| Max. P | 0.969189 |

| Location | 20,640,525 – 20,640,618 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20640525 93 - 22224390 UGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA---AUUUCCCUC-U-CUCGACCA-CUUUUAUUCGACCCG--------------------- ........((((((((((((((..(((((.............))))))))))))))))))---)........-.-.((((...-.......))))....--------------------- ( -20.92) >DroPse_CAF1 46699 92 - 1 UGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGCUGGGCC-AGGCUCCCUUCCAUG-G-CU----CUGGGUGUACUGUACCCC--------------------- ...((((....))))...((((..(((((.............)))))))))(((((-(((.......)).))-)-))----).(((((.....))))).--------------------- ( -22.42) >DroSec_CAF1 44789 94 - 1 UGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA---AUUUCCCUCGU-CCCACCCA-CUUUUAUUCGACCCC--------------------- .(((....((((((((((((((..(((((.............))))))))))))))))))---)........))-).......-...............--------------------- ( -20.32) >DroEre_CAF1 45555 94 - 1 UGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA---AUUUCCCUC-UCCUCGCCCA-CUUUUAUUCGACCCG--------------------- ........((((((((((((((..(((((.............))))))))))))))))))---)........-..........-...............--------------------- ( -19.92) >DroYak_CAF1 46534 93 - 1 UGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA---AUUUCCCUC-U-CUCGCCCA-CUUUUAUUCGACCCG--------------------- ........((((((((((((((..(((((.............))))))))))))))))))---)........-.-........-...............--------------------- ( -19.92) >DroAna_CAF1 6314 109 - 1 UGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA--AAUUUCCCUC-U-------CG-CUUACAUUCUCCCUCUGGCUUUUCAGUUUCAGCCCG ......((((((((((((((((..(((((.............))))))))))))))))))--))).......-.-------..-................((((..........)))).. ( -24.62) >consensus UGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA___AUUUCCCUC_U_CUCGCCCA_CUUUUAUUCGACCCC_____________________ .........(((((((((((((..(((((.............))))))))))))))))))............................................................ (-17.80 = -17.72 + -0.08)

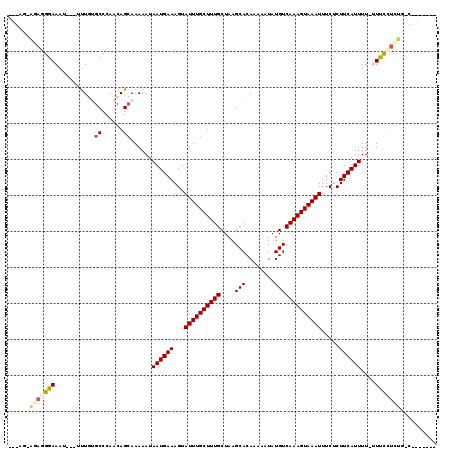
| Location | 20,640,543 – 20,640,657 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20640543 114 + 22224390 UCGAG-AGAGGGAAAU---UUUGUGCCCAACAGCAAAAAUAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU-UUCCCUCUGUCCGACCUG ..((.-((((((((((---(((((........))))))..((((((((.((((((((((....(((........)))))))))))))..)).)))))).)-)))))))).))....... ( -31.90) >DroPse_CAF1 46717 103 + 1 ---AG-CCAUGGAAGGGAGCCU-GGCCCAGCAGCAAAAAUAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCACUUCAUUUU-CUUCUUCU---------- ---..-....(((((..(((.(-(......)))).......(((((.((((((((((((....(((........)))))))))))))...))))))))..-)))))...---------- ( -18.50) >DroEre_CAF1 45573 116 + 1 GCGAGGAGAGGGAAAU---UUUGUGCCCAACAGCAAAAAUAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUUUUUUCCUCUGCCCGACCUG .((.((((((((((((---(((((........))))))..((((((((.((((((((((....(((........)))))))))))))..)).))))))..))))))))).))))..... ( -30.20) >DroYak_CAF1 46552 107 + 1 GCGAG-AGAGGGAAAU---UUUGUGCCCAACAGCAAAAAUAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU-UUCCCUCUGCC------- .....-((((((((((---(((((........))))))..((((((((.((((((((((....(((........)))))))))))))..)).)))))).)-))))))))...------- ( -28.60) >DroAna_CAF1 6352 103 + 1 ------AGAGGGAAAUU--UUUGUGCCCAACAGCAAAAAUAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU-UUUUUGUUUUC------- ------...(((.....--......)))...((((((((.((((((((.((((((((((....(((........)))))))))))))..)).))))))..-))))))))...------- ( -21.20) >DroPer_CAF1 46040 103 + 1 ---AG-CCAUGGAAAGGAGCCU-GGCCCAACAGGAAAAAUAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCACUUCAUUUU-CUUCUUCG---------- ---..-.....(((.(((((((-(......))))......((((((.((((((((((((....(((........)))))))))))))...))))))))..-))))))).---------- ( -20.80) >consensus ___AG_AGAGGGAAAU___UUUGUGCCCAACAGCAAAAAUAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU_UUUCCUCUG_C_______ ......(((((((...........((......))......((((((...((((((((((....(((........))))))))))))).....))))))....))))))).......... (-14.56 = -14.98 + 0.42)

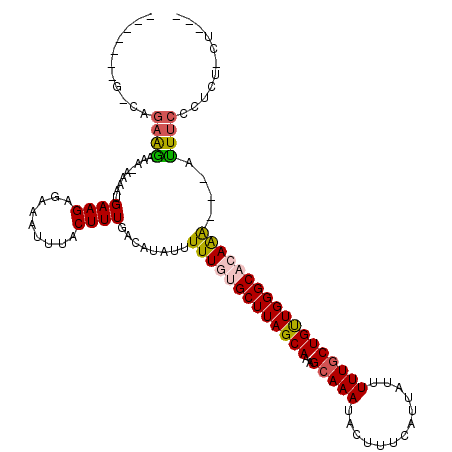
| Location | 20,640,543 – 20,640,657 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -15.63 |
| Energy contribution | -15.69 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20640543 114 - 22224390 CAGGUCGGACAGAGGGAA-AAAAUGAAGAGAAAUUUACUUUGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA---AUUUCCCUCU-CUCGA .......((.((((((((-(..(((.((((.......))))..)))..((((((((((((((..(((((.............))))))))))))))))))---))))))))))-.)).. ( -36.82) >DroPse_CAF1 46717 103 - 1 ----------AGAAGAAG-AAAAUGAAGUGAAAUUUACUUUGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGCUGGGCC-AGGCUCCCUUCCAUGG-CU--- ----------....((((-.....(((((((...)))))))........((((.((((((((..(((((.............))))))))))))))-)))....)))).....-..--- ( -22.02) >DroEre_CAF1 45573 116 - 1 CAGGUCGGGCAGAGGAAAAAAAAUGAAGAGAAAUUUACUUUGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA---AUUUCCCUCUCCUCGC .....((((.(((((.......(((.((((.......))))..)))..((((((((((((((..(((((.............))))))))))))))))))---)....))))).)))). ( -33.12) >DroYak_CAF1 46552 107 - 1 -------GGCAGAGGGAA-AAAAUGAAGAGAAAUUUACUUUGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA---AUUUCCCUCU-CUCGC -------((.((((((((-(..(((.((((.......))))..)))..((((((((((((((..(((((.............))))))))))))))))))---))))))))))-.)).. ( -35.92) >DroAna_CAF1 6352 103 - 1 -------GAAAACAAAAA-AAAAUGAAGAGAAAUUUACUUUGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA--AAUUUCCCUCU------ -------((((.......-...(((.((((.......))))..))).(((((((((((((((..(((((.............))))))))))))))))))--)))))).....------ ( -23.02) >DroPer_CAF1 46040 103 - 1 ----------CGAAGAAG-AAAAUGAAGUGAAAUUUACUUUGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUCCUGUUGGGCC-AGGCUCCUUUCCAUGG-CU--- ----------(((.((((-((((((((((...((((.(((((.((((....)))).....))))).)))))).)))))).))))))...)))((((-(((.......)).)))-))--- ( -20.50) >consensus _______G_CAGAAGAAA_AAAAUGAAGAGAAAUUUACUUUGACAUAUUUUUGUGCUUAGCAAAGCAAAUACUUUCAUUAUUUUUGCUGUUGGGCACAAA___AUUUCCCUCU_CU___ ...........((((.........((((.........))))........(((((((((((((..(((((.............))))))))))))))))))....))))........... (-15.63 = -15.69 + 0.06)

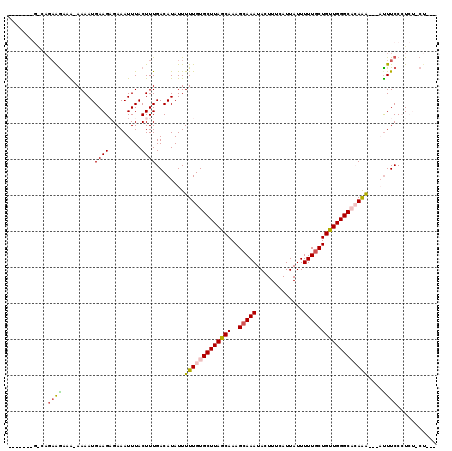
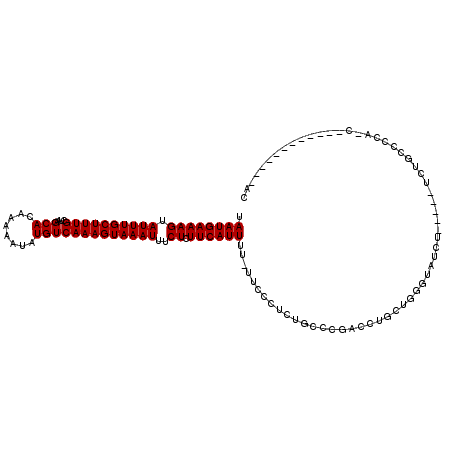
| Location | 20,640,578 – 20,640,697 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20640578 119 + 22224390 UAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU-UUCCCUCUGUCCGACCUGCUGGGUAUCUGGGUAUCUUCACCAUCCUCCCUCUCCCCAC .((((((((.((((((((((....(((........)))))))))))))..)).))))))..-...................((((.....(((...............)))....)))). ( -19.56) >DroSec_CAF1 44843 108 + 1 UAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU-UUCCCUCUGACCGACCUGCUGGGUAUCU-----UCUGCACCACCCACCCCCU------ .((((((((.((((((((((....(((........)))))))))))))..)).))))))..-...................(((((....-----........)))))......------ ( -16.40) >DroEre_CAF1 45609 100 + 1 UAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUUUUUUCCUCUGCCCGACCUGCUGGGUAUCU-----UCUUCCCC---------------AC .((((((((.((((((((((....(((........)))))))))))))..)).))))))..........((((((......))))))...-----........---------------.. ( -18.60) >DroYak_CAF1 46587 91 + 1 UAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU-UUCCCUCUGCC--------UGGGUAUCU-----UCUGCCCC---------------AC .((((((((.((((((((((....(((........)))))))))))))..)).))))))..-...........--------.(((((...-----..))))).---------------.. ( -17.40) >DroAna_CAF1 6383 95 + 1 UAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU-UUUUUGUUUUC--------UUCUUCUUC-----UCUGCCCAACCG-----------AC .((((((((.((((((((((....(((........)))))))))))))..)).))))))..-...........--------.........-----............-----------.. ( -11.80) >consensus UAAUGAAAGUAUUUGCUUUGCUAAGCACAAAAAUAUGUCAAAGUAAAUUUCUCUUCAUUUU_UUCCCUCUGCCCGACCUGCUGGGUAUCU_____UCUGCCCCA_C____________AC .((((((((.((((((((((....(((........)))))))))))))..)).))))))............................................................. (-11.80 = -11.80 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:32 2006