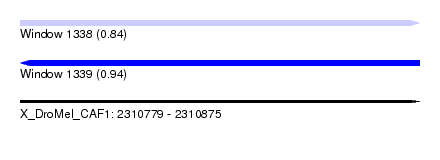
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,310,779 – 2,310,875 |
| Length | 96 |
| Max. P | 0.943946 |

| Location | 2,310,779 – 2,310,875 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
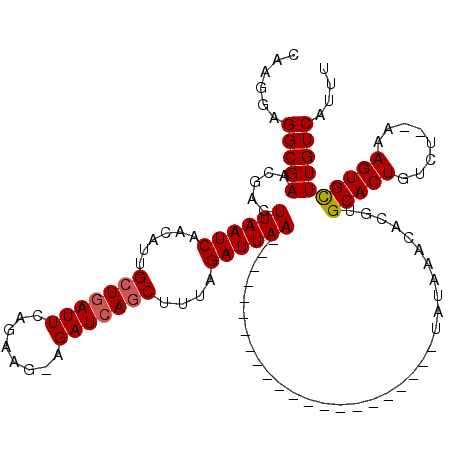
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2310779 96 + 22224390 AAAUGACAAGCACUUU--AGACAGUGCAUGUGUUUAUA----------------------UUAAUCUAAAGAUGAUCUUCUUCUGAAUUAGCAAUGUUGAUUAAGUCGUUCGCCUCCUUG .((((((..(((((..--....)))))...........----------------------....((..((((......))))..))((((((...))))))...)))))).......... ( -15.60) >DroSec_CAF1 8723 95 + 1 AAAUGACAAGCACUUU--GGACAGUGCACGUGUUUAUA----------------------UUAAUCCAAAGCUGAUCU-CUUCUGAAUCAGCAAUGUUGAUUAAGUCGUUCGCCUCCUUG .((((((..(((((..--....)))))...........----------------------((((((((..((((((.(-(....))))))))..))..)))))))))))).......... ( -23.40) >DroSim_CAF1 9544 94 + 1 AAAUGACAAGCACUUU--GGACAGUGCACGUGUUUAUA----------------------UUAAUCCAAAGCUGAUCU-C-UCUGAAUCAGCAAUGUUGAUUAAGUCGUUCGCCUCCUUG .((((((..(((((..--....)))))...........----------------------((((((((..(((((((.-.-...).))))))..))..)))))))))))).......... ( -22.50) >DroEre_CAF1 10013 117 + 1 AAAUGACAAACACUUU--AGACAGUGCGAGUGUUUGUAUGUAUGUAUAUACAUACUCGUAUUAAUCUAAAGCUGAUCU-CUUCUGAAUCAGCAAGGCUGAUUAAGUCGUUCGCCUGCUUG .((((((...((((((--(((.(((((((((((.((((((....)))))).)))))))))))..))))))).))....-......(((((((...)))))))..)))))).......... ( -36.30) >DroYak_CAF1 9726 119 + 1 AAAUGACAAGCACUUUGGAGACAGUGCCAGUGUUUAUAUGCUUGUAUAUUCAUACUCGUCUUAAUCUAAAGCUGAUCU-CUUCUGAAUCAGCAAUGCUGAUUAAGUCGUUCGCCUGUCUG ..................((((((.((.(((((..(((((....)))))..)))))((.(((((((....((((((.(-(....))))))))......))))))).))...)))))))). ( -32.70) >consensus AAAUGACAAGCACUUU__AGACAGUGCAAGUGUUUAUA______________________UUAAUCUAAAGCUGAUCU_CUUCUGAAUCAGCAAUGUUGAUUAAGUCGUUCGCCUCCUUG .((((((..(((((........))))).................................((((((....(((((((.......).))))))......)))))))))))).......... (-18.28 = -18.52 + 0.24)

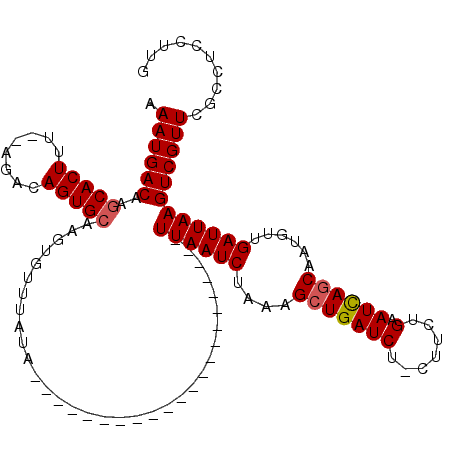
| Location | 2,310,779 – 2,310,875 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2310779 96 - 22224390 CAAGGAGGCGAACGACUUAAUCAACAUUGCUAAUUCAGAAGAAGAUCAUCUUUAGAUUAA----------------------UAUAAACACAUGCACUGUCU--AAAGUGCUUGUCAUUU ......(((((.....((((((.....((......))(((((......))))).))))))----------------------...........(((((....--..)))))))))).... ( -15.30) >DroSec_CAF1 8723 95 - 1 CAAGGAGGCGAACGACUUAAUCAACAUUGCUGAUUCAGAAG-AGAUCAGCUUUGGAUUAA----------------------UAUAAACACGUGCACUGUCC--AAAGUGCUUGUCAUUU ......(((((.....((((((..((..(((((((......-.)))))))..))))))))----------------------...........(((((....--..)))))))))).... ( -24.10) >DroSim_CAF1 9544 94 - 1 CAAGGAGGCGAACGACUUAAUCAACAUUGCUGAUUCAGA-G-AGAUCAGCUUUGGAUUAA----------------------UAUAAACACGUGCACUGUCC--AAAGUGCUUGUCAUUU ......(((((.....((((((..((..(((((((....-.-.)))))))..))))))))----------------------...........(((((....--..)))))))))).... ( -24.60) >DroEre_CAF1 10013 117 - 1 CAAGCAGGCGAACGACUUAAUCAGCCUUGCUGAUUCAGAAG-AGAUCAGCUUUAGAUUAAUACGAGUAUGUAUAUACAUACAUACAAACACUCGCACUGUCU--AAAGUGUUUGUCAUUU ......(((((((..((.(((((((...))))))).))...-......(((((((((.....(((((.(((((........)))))...)))))....))))--)))))))))))).... ( -34.70) >DroYak_CAF1 9726 119 - 1 CAGACAGGCGAACGACUUAAUCAGCAUUGCUGAUUCAGAAG-AGAUCAGCUUUAGAUUAAGACGAGUAUGAAUAUACAAGCAUAUAAACACUGGCACUGUCUCCAAAGUGCUUGUCAUUU ..((((.((...((.(((((((......(((((((......-.)))))))....))))))).)).((((....))))..))...........((((((........)))))))))).... ( -31.30) >consensus CAAGGAGGCGAACGACUUAAUCAACAUUGCUGAUUCAGAAG_AGAUCAGCUUUAGAUUAA______________________UAUAAACACGUGCACUGUCU__AAAGUGCUUGUCAUUU ......(((((.....((((((......(((((((........)))))))....)))))).................................(((((........)))))))))).... (-19.66 = -19.90 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:46 2006