| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,373,897 – 20,374,055 |
| Length | 158 |
| Max. P | 0.625634 |

| Location | 20,373,897 – 20,373,999 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -15.89 |
| Energy contribution | -15.78 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20373897 102 - 22224390 UCGUUUUAACACACUUGAGGCCAAAUACUUGGCCAAGUUUUGCCAAAGUUUGCAAACCAAAAUUUCACCGAAAAUGUGACACCAAC-----------GAACCAACCAAACGAA (((((....((((.....((((((....))))))..(.(((((........))))).)................)))).....)))-----------)).............. ( -19.80) >DroVir_CAF1 42655 112 - 1 CCGCUCUGUCGCACUUGAGGCCAAAUACUUGGCCAAGUUUUGCCAAAGUUUGCA-ACCAAAAUUUCACCGAAAAUGUGACACCAACAACCCACCAACGAACCAACCAGACCAA ......(((((((.....((((((....))))))...(((((...((((((...-....))))))...))))).)))))))................................ ( -20.80) >DroSim_CAF1 22397 101 - 1 UCGUUUUAACACACUUGAGGCCAAAUACUUGGCCCAGUUUUGCCAAAGUUUGCAAACCAA-AUUUCACCGAAAAUGUGAACACCAC-----------GAACCAACCANNNNNN ((((.....((((.(((.((((((....)))))))))(((((...(((((((.....)))-))))...))))).))))......))-----------)).............. ( -20.10) >DroYak_CAF1 11146 102 - 1 UCGCUUUAACACACUUGAGGCCAAAUACUUGGCCAAGUUUUGCCAAAGUUUGCAAACCAAAAUUUCACCGAAAAUGUGACACCAAC-----------GAACCAACCAAACGAA (((......((((.....((((((....))))))..(.(((((........))))).)................)))).......)-----------)).............. ( -17.02) >DroAna_CAF1 42971 102 - 1 UCGCUCCGGCACACUUGAGGCUAAAUACUUGGCCAAGUUUUGCCAAAGUUUGCAAACCAAAAUUUUACCGAAAAUGUGACACCAAC-----------GAACCAACCAAAGGAA ....(((((((.((((..((((((....))))))))))..))))...(((((.........(((((....)))))..........)-----------))))........))). ( -20.31) >DroPer_CAF1 42946 102 - 1 UCGUUCUAACACACUUGAGGCCAAAUACUUGGCCAAGUUUUGCCAAAGUUUGCAAACCAAAAUUUCACCGAAAAUGUGACACCAAC-----------GAACCAACCAAACGAA (((((....((((.....((((((....))))))..(.(((((........))))).)................)))).....)))-----------)).............. ( -19.80) >consensus UCGCUCUAACACACUUGAGGCCAAAUACUUGGCCAAGUUUUGCCAAAGUUUGCAAACCAAAAUUUCACCGAAAAUGUGACACCAAC___________GAACCAACCAAACGAA .........((((.....((((((....))))))..(.(((((........))))).)................))))................................... (-15.89 = -15.78 + -0.11)
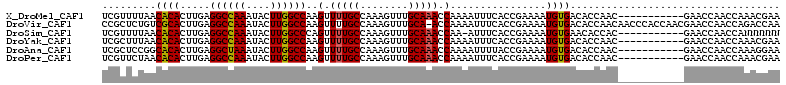
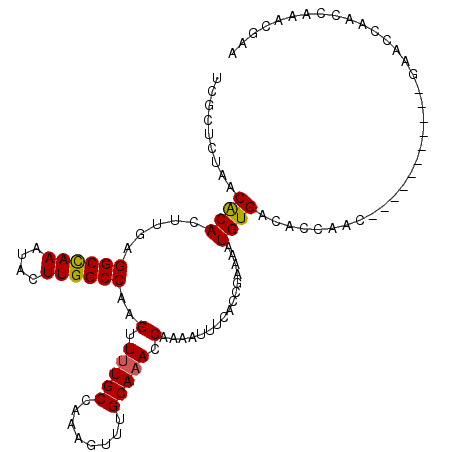
| Location | 20,373,962 – 20,374,055 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -25.39 |
| Energy contribution | -26.75 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20373962 93 - 22224390 UCAAGCACCCAGCUAGCCGUCUCUUUCGCCGGCGGACUGGCUGGCUUGUCUCCGUCUCGUUUUAACACACUUGAGGCCAAAUACUUGGCCAAG (((((...(((((((((((((.........))))).))))))))..............((....))...)))))((((((....))))))... ( -33.90) >DroSec_CAF1 56513 93 - 1 UCAAGCACCCAGCUAGCCGUCUCUUUCGCCGGCGGACUGGCUGGCCUGUCUCCGUCUCGUUUUAACACACUUGAGGCCAAAUACUUGGCCAAG (((((...(((((((((((((.........))))).))))))))..............((....))...)))))((((((....))))))... ( -33.90) >DroSim_CAF1 22461 93 - 1 UCAAGCACCCAGCUAGCCGUCUCUUUCGCCGGCGGACUGGCUGGCUUGUCUCCGUCUCGUUUUAACACACUUGAGGCCAAAUACUUGGCCCAG .(((((...((((((((((((.........))))).)))))))))))).......................((.((((((....)))))))). ( -33.10) >DroEre_CAF1 20833 93 - 1 UCGCGCACCCAGCUAGCCGUCUCUCUCGCCGGCGUACUGGCUGGCUUGUCUCCGUCUCGUUUUAACACACUUGAGGCCAAAUACUUGGCCAAG ..((.((.((((...((((.(......).))))...)))).)))).............................((((((....))))))... ( -26.50) >DroYak_CAF1 11211 93 - 1 UCGCGCACCCAGCUAGCCGUCUCUUUCGCCGGCGGACUGGCUGGCUUGUCUCCGUCUCGCUUUAACACACUUGAGGCCAAAUACUUGGCCAAG ..(((.(((((((((((((((.........))))).)))))))).........))..)))..............((((((....))))))... ( -31.90) >DroMoj_CAF1 51876 87 - 1 UCAG--UCGCGCCCAGACGACUCUGUCGU----ACUCUGGCUUGGCUGCCUCUGUCCCGCUCUGACGCACUUGAGGCCAAAUACUUGGCCAAG .(((--(((.(((.(((((((...)))))----.))..))).))))))..........((......))......((((((....))))))... ( -28.10) >consensus UCAAGCACCCAGCUAGCCGUCUCUUUCGCCGGCGGACUGGCUGGCUUGUCUCCGUCUCGUUUUAACACACUUGAGGCCAAAUACUUGGCCAAG (((((...(((((((((((((.........))))).))))))))..............((......)).)))))((((((....))))))... (-25.39 = -26.75 + 1.36)

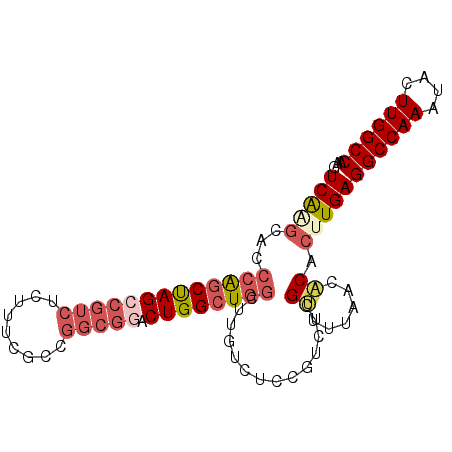
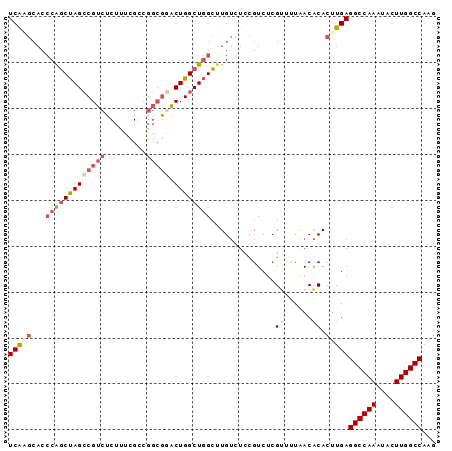
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:16 2006