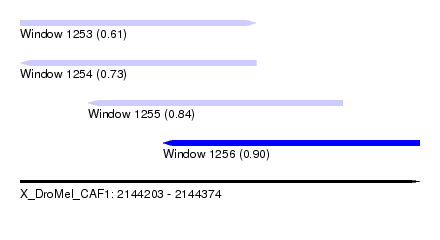
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,144,203 – 2,144,374 |
| Length | 171 |
| Max. P | 0.902590 |

| Location | 2,144,203 – 2,144,304 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.94 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2144203 101 + 22224390 CUUUUAGAAUGCAA-----------CAACGCAAUGCAAAUCAAUCGAUGAGCAAAUUUGAUU--------GCCGCAUUGCAGAACGAUAAGCGCAUGCAAAAGCAUGGCCGACAAUCAGC ......((.(((..-----------....(((((((....(((((((.........))))))--------)..)))))))..........((.(((((....))))))).).)).))... ( -26.10) >DroSec_CAF1 24633 101 + 1 CUUUAAGAAUGCAA-----------CAACGCAAUGCAAAUCAAUCGAUGAGCAAUUUUUAUU--------GUCGCAUUGCAGAACGAUAAGCGCAUGCAAAAGCAUGGCCGACAAUCAGC ......((.(((..-----------....(((((((..(((....)))((.((((....)))--------))))))))))..........((.(((((....))))))).).)).))... ( -24.30) >DroSim_CAF1 22048 101 + 1 CUUUAAGAAUGCAA-----------CAACGCAAUGCAAAUCAAUCGAUGAGCAAAUUUGAUU--------GCCGCAUUGCAGAACGAUAAGCGCAUGCAAAAGCAUGGCCGACAAUCAGC ......((.(((..-----------....(((((((....(((((((.........))))))--------)..)))))))..........((.(((((....))))))).).)).))... ( -26.10) >DroEre_CAF1 25479 103 + 1 C-UUAAGAAUGCAAGUGCAAUAACG--------UGCAAAUCAAUCGAUGAGCAAAUUUGAUU--------ACCGCACUGCAGAACGAUAAGCGCAUGCAAAAGCAUGAUCGACAAUCAGC .-.......((((.(((((((....--------(((..(((....)))..)))......)))--------...))))))))....(((...(((((((....)))))..))...)))... ( -21.80) >DroYak_CAF1 27335 119 + 1 C-UUAAGAAUGCAAGUGCAAUAACGCAAUGCAAUGCAAAUCAAUCGAUGAGCAAAUUUGAUUGCCGGAUUGCCGCAUUGCAGAACGAUAAGCGCAUGCAAAAGCAUGGUCGACAAUCAGC .-.......((((.((((.....((...((((((((....(((((((.........)))))))..((....))))))))))...))....)))).))))...((.((((.....)))))) ( -31.70) >consensus CUUUAAGAAUGCAA___________CAACGCAAUGCAAAUCAAUCGAUGAGCAAAUUUGAUU________GCCGCAUUGCAGAACGAUAAGCGCAUGCAAAAGCAUGGCCGACAAUCAGC ..........((.................(((((((.((((((.............))))))...........))))))).....(((...(((((((....)))))..))...))).)) (-15.94 = -16.94 + 1.00)

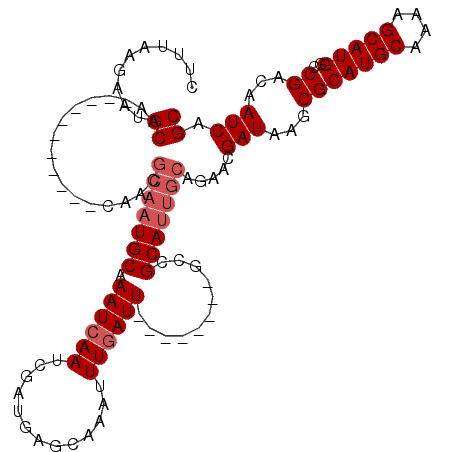
| Location | 2,144,203 – 2,144,304 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.86 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2144203 101 - 22224390 GCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGC--------AAUCAAAUUUGCUCAUCGAUUGAUUUGCAUUGCGUUG-----------UUGCAUUCUAAAAG ((.((.((..((((((((....))))).)))...)).)).))((((((((--------((((((...(((..(((....)))..)))))).))))-----------)))))))....... ( -30.60) >DroSec_CAF1 24633 101 - 1 GCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGAC--------AAUAAAAAUUGCUCAUCGAUUGAUUUGCAUUGCGUUG-----------UUGCAUUCUUAAAG ((.((.((..((((((((....))))).)))...)).)).))((((((((--------(((......(((..(((....)))..)))....))))-----------)))))))....... ( -30.40) >DroSim_CAF1 22048 101 - 1 GCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGC--------AAUCAAAUUUGCUCAUCGAUUGAUUUGCAUUGCGUUG-----------UUGCAUUCUUAAAG ((.((.((..((((((((....))))).)))...)).)).))((((((((--------((((((...(((..(((....)))..)))))).))))-----------)))))))....... ( -30.60) >DroEre_CAF1 25479 103 - 1 GCUGAUUGUCGAUCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAGUGCGGU--------AAUCAAAUUUGCUCAUCGAUUGAUUUGCA--------CGUUAUUGCACUUGCAUUCUUAA-G ...(((...((..(((((....)))))))...)))....(((((((((((--------(((((((((..(.....)...))))))..--------.)))))))))).)))).......-. ( -25.80) >DroYak_CAF1 27335 119 - 1 GCUGAUUGUCGACCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGCAAUCCGGCAAUCAAAUUUGCUCAUCGAUUGAUUUGCAUUGCAUUGCGUUAUUGCACUUGCAUUCUUAA-G ...((.((.(((.(((((....))))).((....(((..(((((((((.(((((.(((((......)))))....)))))...)))))))))..))).....))..))))).))....-. ( -36.20) >consensus GCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGC________AAUCAAAUUUGCUCAUCGAUUGAUUUGCAUUGCGUUG___________UUGCAUUCUUAAAG ((.((.((..((((((((....))))).)))...)).)).))(((((((............(((...(((..(((....)))..))).....)))...........)))))))....... (-18.14 = -18.86 + 0.72)


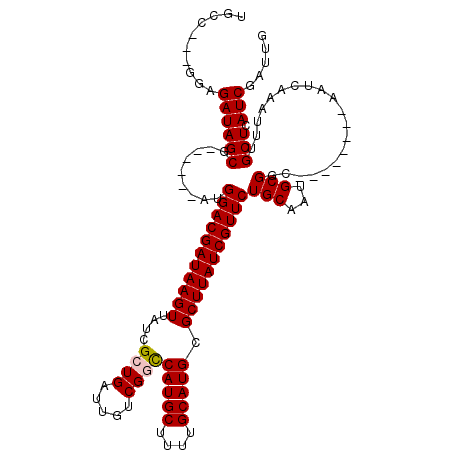
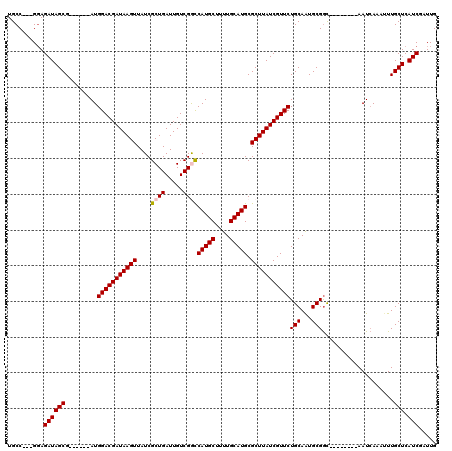
| Location | 2,144,232 – 2,144,341 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.71 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -26.78 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2144232 109 - 22224390 UGCC---GGAGAUAGCGAUGGCGAUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGC--------AAUCAAAUUUGCUCAUCGAUUG ....---........(((((((((.(((((((((((....((((.....))))(((((....))))).)))))))))))(((......))--------).......)))).))))).... ( -37.00) >DroSec_CAF1 24662 103 - 1 UGCC---GGAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGAC--------AAUAAAAAUUGCUCAUCGAUUG ....---...(((((((------(((((((((((((....((((.....))))(((((....))))).)))))))))))(((...)))..--------.......)))))).)))..... ( -32.10) >DroSim_CAF1 22077 103 - 1 UGCC---GGAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGC--------AAUCAAAUUUGCUCAUCGAUUG ((((---(.(....(((------..(((((((((((....((((.....))))(((((....))))).))))))))))))))..).))))--------)..................... ( -34.70) >DroEre_CAF1 25510 103 - 1 UGCC---GUAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGAUCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAGUGCGGU--------AAUCAAAUUUGCUCAUCGAUUG ((((---(((....(((------..(((((((((((.(((((.....).))))(((((....))))).))))))))))))))..))))))--------)..................... ( -31.10) >DroYak_CAF1 27374 114 - 1 UGCCGCCGGAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGACCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGCAAUCCGGCAAUCAAAUUUGCUCAUCGAUUG .((((((((((((((((------...((((((.(((....))).))))))...(((((....)))))))).))))..))))....)))))((((.(((((......)))))....)))). ( -37.20) >consensus UGCC___GGAGAUAGCG______AUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCUGCAAUGCGGC________AAUCAAAUUUGCUCAUCGAUUG ..........((((((.........(((((((((((....((((.....))))(((((....))))).)))))))))))(((...)))....................))).)))..... (-26.78 = -27.02 + 0.24)

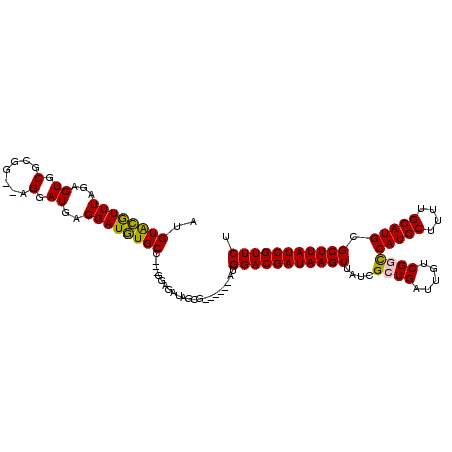
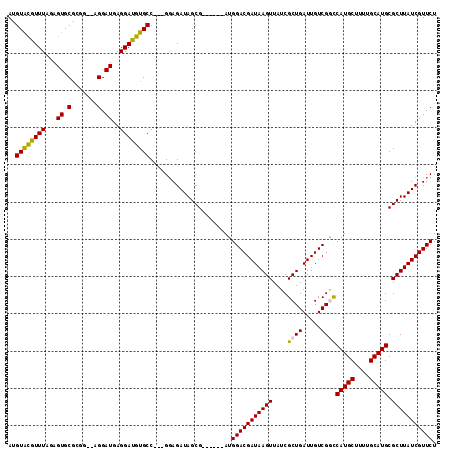
| Location | 2,144,264 – 2,144,374 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -28.60 |
| Energy contribution | -27.96 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2144264 110 - 22224390 AUGUACGUUUAGAGUGCACGG--AGGAUGAGGAUGUGCC---GGAGAUAGCGAUGGCGAUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCU .(((((.......))))).((--(.((((((.....(((---(..((((((((((((............))))))))).)))..))))(((((....)))))..)))))).))). ( -40.90) >DroSec_CAF1 24694 104 - 1 AUGUACGUUUAGAGUGCGCGG--AGGAUGAGGAUGUGCC---GGAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCU ..((((.......))))..((--(.((((((.....(((---(..(((((((------(((.((.....))))))))).)))..))))(((((....)))))..)))))).))). ( -38.00) >DroSim_CAF1 22109 104 - 1 AUGUACGUUUAGAGUGCGCGG--AGGAUGAGGAUGUGCC---GGAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCU ..((((.......))))..((--(.((((((.....(((---(..(((((((------(((.((.....))))))))).)))..))))(((((....)))))..)))))).))). ( -38.00) >DroEre_CAF1 25542 106 - 1 AUGUGUGUUUAGAGUGCGAGGGGGGGAUGAGGAUAUGCC---GUAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGAUCAUGCUUUUGCAUGCGCUUAUCGUUCU ...........(((((((.....((.(((....))).))---(((((.((((------((.((((((.(((....))).)))))).)))..)))))))).)))))))........ ( -28.40) >DroYak_CAF1 27414 107 - 1 AUGUGUAUUUAGAGUGCGAGC--AGGAUGAGGAUAUGCCGCCGGAGAUAGCG------AUGGACGAUAAGUUAUCGCUGAUUGUCGACCAUGCUUUUGCAUGCGCUUAUCGUUCU .(((.(((((...(.(((.((--(...........)))))))..))))))))------..(((((((((((..((((.....).))).(((((....))))).))))))))))). ( -30.70) >consensus AUGUACGUUUAGAGUGCGCGG__AGGAUGAGGAUGUGCC___GGAGAUAGCG______AUGGACGAUAAGUUAUCGCUGAUUGUCGGCCAUGCUUUUGCAUGCGCUUAUCGUUCU ..((((((((...((.(.......).))..))))))))......................(((((((((((....((((.....))))(((((....))))).))))))))))). (-28.60 = -27.96 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:30 2006