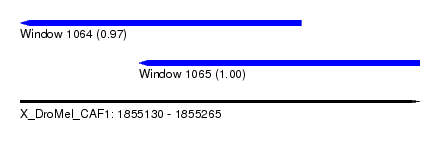
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,855,130 – 1,855,265 |
| Length | 135 |
| Max. P | 0.995111 |

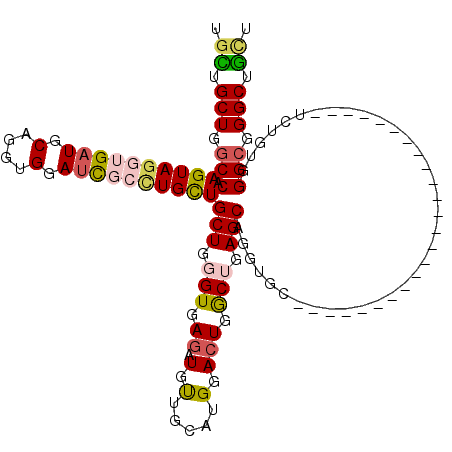
| Location | 1,855,130 – 1,855,225 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -44.86 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1855130 95 - 22224390 GCUGUGUGAGAUGUUGCAUGGACUGACUGAGCAGGUGC------------------------UCUGUGGGCGGGCUGCUCUUGCGCAGCGUCAGCGGCUGUGUGAGUUGCUGGUGAAGA ((((((..(((.((.((.(.(.((.((.((((....))------------------------)).)).))).))).)))))..)))))).(((((((((.....)))))))))...... ( -43.00) >DroPse_CAF1 89827 119 - 1 GCUGGGUGAGGUGCUGCAUGGACUGGCUGAGCAGGUGCUGCGGACUCUGGCCCAGCUGCAGCUCCAUUGGCGGGCUGUUCUUCCGCAGGGUCAGCGGCUGUGUCAGUUGCUGGUGCAGG (((((((.((((.(((((.(.(((.((...)).))).)))))))).)).)))))))((((...((....(((((.......)))))..))((((((((((...)))))))))))))).. ( -54.20) >DroSec_CAF1 55996 95 - 1 GCUGGGUGAGAUGUUGCAUGGACUGGCUGAGCAGGUGC------------------------UCUGUGGGCGGGCUGCUCUUGCGCAGCGUCAGCGGCUGUGUGAGUUGCUGGUGAAGA ((((.(..(((.((.((.(.(.((.((.((((....))------------------------)).)).))).))).)))))..).)))).(((((((((.....)))))))))...... ( -41.10) >DroSim_CAF1 48952 95 - 1 GCUGGGUGAGAUGUUGCAUGGACUGGCUGAGCAGGUGC------------------------UCUGUGGGCGGGCUGCUCUUGCGCAGCGUCAGCGGCUGUGUGAGUUGCUGGUGAAGA ((((.(..(((.((.((.(.(.((.((.((((....))------------------------)).)).))).))).)))))..).)))).(((((((((.....)))))))))...... ( -41.10) >DroWil_CAF1 89572 119 - 1 GCUGCGUCAAAUGCUGCAUGGACUGGCUAAGCAAAUGCUGUGGACUCUGGCCCAAUUGCAGAUCUGUGGGCGGGCUAGUCUUUCGCAAGGUCAGCGGUUGGGUUAGCUGCUGAUGCAAA ..(((((((...(((((((.(.((((((.(((....)))(((((..(((((((..(..((....))..)..)))))))...)))))..))))))).))...).))))...))))))).. ( -44.90) >consensus GCUGGGUGAGAUGUUGCAUGGACUGGCUGAGCAGGUGC________________________UCUGUGGGCGGGCUGCUCUUGCGCAGCGUCAGCGGCUGUGUGAGUUGCUGGUGAAGA ((((.(..(((.((.((.....(((......))).............................(((....))))).)))))..).)))).(((((((((.....)))))))))...... (-25.08 = -25.64 + 0.56)

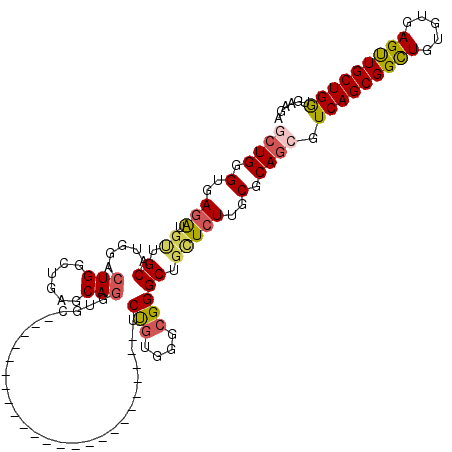
| Location | 1,855,170 – 1,855,265 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -26.32 |
| Energy contribution | -27.16 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1855170 95 - 22224390 UGCUGCUGGCCAAGUAGGUGAUGCAGGUGGAUCGCCUGCUGCUGUGUGAGAUGUUGCAUGGACUGACUGAGCAGGUGC------------------------UCUGUGGGCGGGCUGCU .((.(((.(((.((((((((((.(....).)))))))))).(((((..(....)..)))))....((.((((....))------------------------)).)).))).))).)). ( -41.80) >DroPse_CAF1 89867 119 - 1 UGUUGCUGGCCGAGUAGGUGAUGCAGGUGGAGCUGUUGCUGCUGGGUGAGGUGCUGCAUGGACUGGCUGAGCAGGUGCUGCGGACUCUGGCCCAGCUGCAGCUCCAUUGGCGGGCUGUU ....(((.(((..(((.....))).(((((((((((....(((((((.((((.(((((.(.(((.((...)).))).)))))))).)).))))))).)))))))))))))).))).... ( -60.90) >DroSec_CAF1 56036 95 - 1 UGCUGCUGGCCAAGUAGGUGAUGCAGGUGGAUCGCCUGCUGCUGGGUGAGAUGUUGCAUGGACUGGCUGAGCAGGUGC------------------------UCUGUGGGCGGGCUGCU .((.(((.(((.((((((((((.(....).))))))))))((..((((........)))...)..)).((((....))------------------------))....))).))).)). ( -39.80) >DroSim_CAF1 48992 95 - 1 UGCUGCUGGCCAAGUAGGUGAUGCAGGUGGAUCGCCUGCUGCUGGGUGAGAUGUUGCAUGGACUGGCUGAGCAGGUGC------------------------UCUGUGGGCGGGCUGCU .((.(((.(((.((((((((((.(....).))))))))))((..((((........)))...)..)).((((....))------------------------))....))).))).)). ( -39.80) >DroWil_CAF1 89612 119 - 1 UGUUGCUGUCCUAGUAAAUGAUGCAAAUGCAUUUGCUGUUGCUGCGUCAAAUGCUGCAUGGACUGGCUAAGCAAAUGCUGUGGACUCUGGCCCAAUUGCAGAUCUGUGGGCGGGCUAGU .(((((((((((((((..(((((((...(((........))))))))))..)))))...)))).)))..(((....)))...))).(((((((..(..((....))..)..))))))). ( -41.70) >consensus UGCUGCUGGCCAAGUAGGUGAUGCAGGUGGAUCGCCUGCUGCUGGGUGAGAUGUUGCAUGGACUGGCUGAGCAGGUGC________________________UCUGUGGGCGGGCUGCU .((.(((.(((.((((((((((.(....).))))))))))(((.(((.((.(.(.....).))).))).)))....................................))).))).)). (-26.32 = -27.16 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:33 2006