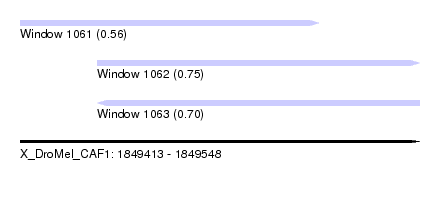
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,849,413 – 1,849,548 |
| Length | 135 |
| Max. P | 0.752113 |

| Location | 1,849,413 – 1,849,514 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -43.35 |
| Consensus MFE | -32.90 |
| Energy contribution | -34.40 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1849413 101 + 22224390 AUCCAAGGCCAGCAGUGUUGUUGCGGCAGCUGCCGAGCUAGAGGCAGCAACAACCACGGCAGCGCUGGCCACGUCCUCCGCCACGGAGGCUUCUGCAGCGG ..((..(((((((...(((((((.((..((((((........)))))).....)).))))))))))))))..(.((((((...))))))).........)) ( -46.50) >DroSec_CAF1 50946 101 + 1 AUCCAAGGCCAGCAGUGUUGUUGCGGCAGCUGCCGAGCUAGAGGCAGCAACAACUACGGCAGCGCUGGCCACGGCCUCGGCCACGGAGGCUUCUGCAGCGG .(((..(((((((..((((((.(..(..((((((........))))))..)..).))))))..)))))))..(((....)))..))).(((.....))).. ( -47.30) >DroEre_CAF1 52276 101 + 1 AUCCAAGGCCAGCAGUGUUGUUGCGGCAACUGCGGAACUAGAGGCAGCAACUGCCACCGGAUCGCUGGCCACGGCCUCGGCCACGGAGGCUUCUGCAGCGG .(((..(((((((.(((((((....))))).)).((.(..(.(((((...))))).)..).)))))))))..(((....)))..))).(((.....))).. ( -42.30) >DroYak_CAF1 42182 86 + 1 AUCCAAGGCCAGCAGUGUUGUUGCGGCAGCUGCCGAUCUAGAGGCAGCCAC---------------GGCCACGGCCUCGGCCACGGAGGCAACUGCAGCGG ..((.(((((.((((.....))))(((.((((((........))))))...---------------.)))..))))).))((.((.((....)).).).)) ( -37.30) >consensus AUCCAAGGCCAGCAGUGUUGUUGCGGCAGCUGCCGAGCUAGAGGCAGCAACAACCACGGCAGCGCUGGCCACGGCCUCGGCCACGGAGGCUUCUGCAGCGG .(((..(((((((..(((((...)))))((((((........))))))...............)))))))..(((....)))..))).((....))..... (-32.90 = -34.40 + 1.50)

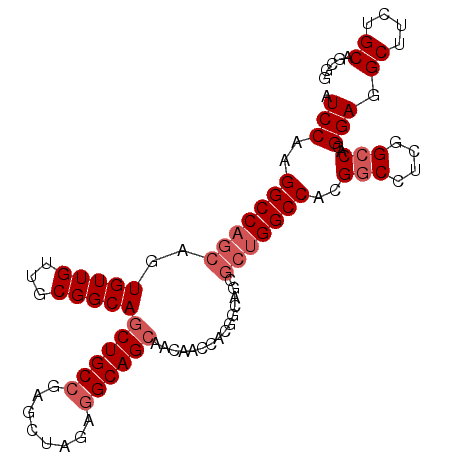
| Location | 1,849,439 – 1,849,548 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -46.35 |
| Consensus MFE | -35.40 |
| Energy contribution | -35.77 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1849439 109 + 22224390 CAGCUGCCGAGCUAGAGGCAGCAACAACCACGGCAGCGCUGGCCACGUCCUCCGCCACGGAGGCUUCUGCAGCGGACAGCUAUCCUGCCGUACAGCAGAACGGCGGCCA ..((((((........)))))).........(((..(((((.....(.((((((...)))))))((((((.((((.(((.....)))))))...)))))))))))))). ( -48.80) >DroSec_CAF1 50972 109 + 1 CAGCUGCCGAGCUAGAGGCAGCAACAACUACGGCAGCGCUGGCCACGGCCUCGGCCACGGAGGCUUCUGCAGCGGACAGCUAUCCCGCCGUACAGCAGAACGGCGGCCA ..(((((((.((.....))((......)).)))))))...((((..(((....)))..(((((((((((...)))).)))).))).(((((........))))))))). ( -46.80) >DroEre_CAF1 52302 109 + 1 CAACUGCGGAACUAGAGGCAGCAACUGCCACCGGAUCGCUGGCCACGGCCUCGGCCACGGAGGCUUCUGCAGCGGACAGCUAUACCGCCGUGCAGCAGAACGGUGGCCA .....((((..(..(.(((((...))))).)..).)))).(((((((((....))).((....((.(((((((((.........))))..))))).))..)))))))). ( -45.30) >DroYak_CAF1 42208 94 + 1 CAGCUGCCGAUCUAGAGGCAGCCAC---------------GGCCACGGCCUCGGCCACGGAGGCAACUGCAGCGGACAGCUAUGCCGCCGUGCAGCAGAAUGGCGGCCA ..(((((((.(((.(((((.((...---------------.))....))))).(((((((.((((...((........))..)))).)))))..))))).))))))).. ( -44.50) >consensus CAGCUGCCGAGCUAGAGGCAGCAACAACCACGGCAGCGCUGGCCACGGCCUCGGCCACGGAGGCUUCUGCAGCGGACAGCUAUCCCGCCGUACAGCAGAACGGCGGCCA ..((((((........))))))..................((((...(((((.......)))))......(((.....))).....(((((........))))))))). (-35.40 = -35.77 + 0.37)

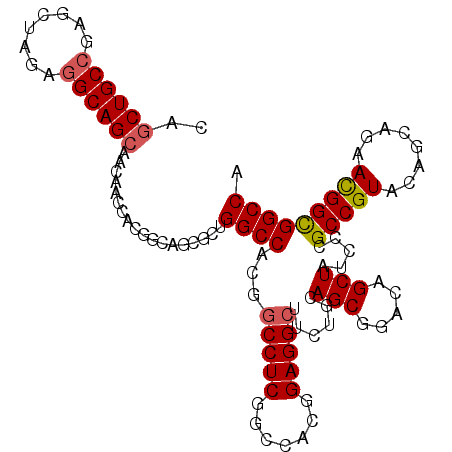
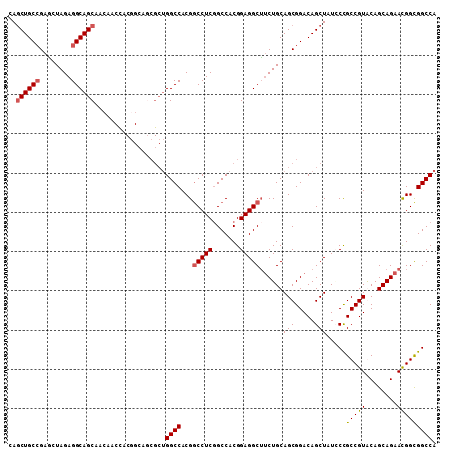
| Location | 1,849,439 – 1,849,548 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -51.30 |
| Consensus MFE | -42.58 |
| Energy contribution | -43.20 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1849439 109 - 22224390 UGGCCGCCGUUCUGCUGUACGGCAGGAUAGCUGUCCGCUGCAGAAGCCUCCGUGGCGGAGGACGUGGCCAGCGCUGCCGUGGUUGUUGCUGCCUCUAGCUCGGCAGCUG (((((((..((((((.(((((((......)))))..)).))))))(((((((...)))))).))))))))..(((((((.(((((..(....)..)))))))))))).. ( -53.30) >DroSec_CAF1 50972 109 - 1 UGGCCGCCGUUCUGCUGUACGGCGGGAUAGCUGUCCGCUGCAGAAGCCUCCGUGGCCGAGGCCGUGGCCAGCGCUGCCGUAGUUGUUGCUGCCUCUAGCUCGGCAGCUG (((((((..((((((.(((((((......)))))..)).))))))(((((.......))))).)))))))..(((((((.(((((..(....)..)))))))))))).. ( -52.80) >DroEre_CAF1 52302 109 - 1 UGGCCACCGUUCUGCUGCACGGCGGUAUAGCUGUCCGCUGCAGAAGCCUCCGUGGCCGAGGCCGUGGCCAGCGAUCCGGUGGCAGUUGCUGCCUCUAGUUCCGCAGUUG (((((((..((((((.(((((((......)))))..)).))))))(((((.......))))).)))))))(((..(..(.(((((...))))).)..)...)))..... ( -50.50) >DroYak_CAF1 42208 94 - 1 UGGCCGCCAUUCUGCUGCACGGCGGCAUAGCUGUCCGCUGCAGUUGCCUCCGUGGCCGAGGCCGUGGCC---------------GUGGCUGCCUCUAGAUCGGCAGCUG .((((((......(((((((((((((...)))).))).)))))).(((((.......))))).))))))---------------..(((((((........))))))). ( -48.60) >consensus UGGCCGCCGUUCUGCUGCACGGCGGGAUAGCUGUCCGCUGCAGAAGCCUCCGUGGCCGAGGCCGUGGCCAGCGCUGCCGUGGUUGUUGCUGCCUCUAGCUCGGCAGCUG (((((((..((((((.(((((((......)))))..)).))))))(((((.......))))).))))))).................((((((........)))))).. (-42.58 = -43.20 + 0.62)

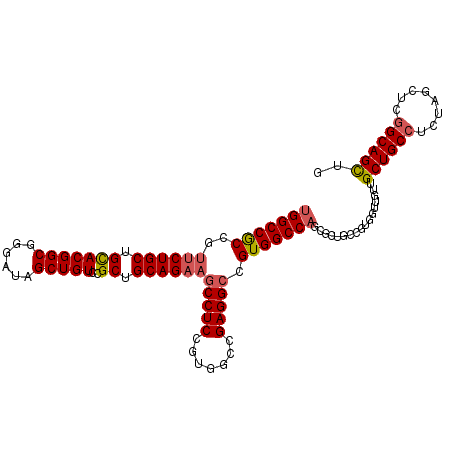
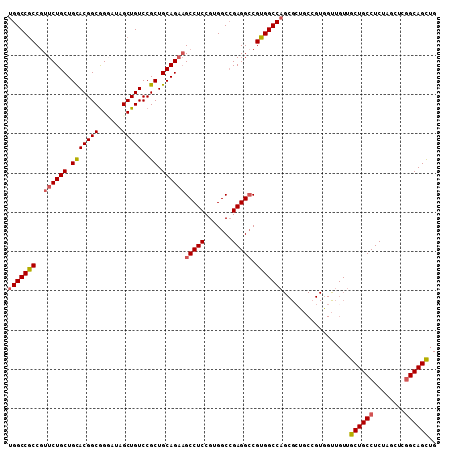
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:31 2006