| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,844,303 – 1,844,398 |
| Length | 95 |
| Max. P | 0.599642 |

| Location | 1,844,303 – 1,844,398 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.23 |
| Mean single sequence MFE | -26.67 |
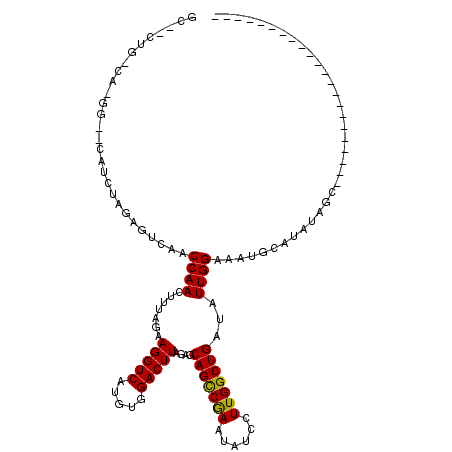
| Consensus MFE | -17.08 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
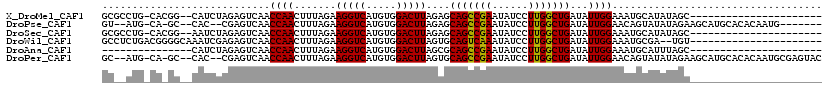
>X_DroMel_CAF1 1844303 95 - 22224390 GCGCCUG-CACGG--CAUCUAGAGUCAACCAACUUUAGAAGGUCAUGUGGACUUAGAGCAGCCGAAUAUCCUUGGCUGAUAUUGGAAAUGCAUAUAGC---------------------- (((.(..-((.((--(.((((((((......))))))))..))).))..).((.....(((((((......))))))).....))...))).......---------------------- ( -28.40) >DroPse_CAF1 77252 105 - 1 GU--AUG-CA-GC--CAC--CGAGUCAACCAACUUUAGAAGGUCAUGUGGACUUAGAGCAGCCGAAUAUCCUUGGCUGAUAUUGGAACAGUAUAUAGAAGCAUGCACACAAUG------- ((--(((-(.-..--..(--..((((.((..((((....))))...)).))))..)..(((((((......)))))))((((((...))))))......))))))........------- ( -27.00) >DroSec_CAF1 45867 95 - 1 GCGCCUG-CACGG--AAUCUAGAGUCAACCAACUUUAGAAGGUCAUGUGGACUUAGAGCAGCCGAAUAUCCUUGGCUGAUAUUGGAAAUGCAUAUAGC---------------------- (((.(..-((.((--..((((((((......))))))))...)).))..).((.....(((((((......))))))).....))...))).......---------------------- ( -25.70) >DroWil_CAF1 75895 96 - 1 GCCUCUGACGGGGCAAAUCGAGAGUCAACCAACUUUAGAAGGUCAUGUGGACUUAGUGCAGUCAAAUAUCCUUGGCUGAUAUUGGAAAUGCGA--UGU---------------------- (((((....)))))..((((.(((((.((..((((....))))...)).)))))(((((((((((......))))))).)))).......)))--)..---------------------- ( -25.50) >DroAna_CAF1 54627 83 - 1 ---------------CAUCUAGAGUCAACCAACUUUAGAAGGUCAUGUGGACUUAGCGCAGCCGAAUAUCCUUGGCUGAUAUUGGAAAUGCAUUUAGC---------------------- ---------------..((((((((......))))))))(((((.....)))))....(((((((......)))))))....................---------------------- ( -21.00) >DroPer_CAF1 79940 112 - 1 GC--AUG-CA-GC--CAC--CGAGUCAACCAACUUUAGAAGGUCAUGUGGACUUAGUGCAGCCGAAUAUCCUUGGCUGAUAUUGGAACAGUAUAUAGAAGCAUGCACACAAUGCGAGUAC ((--(((-(.-..--(((--..((((.((..((((....))))...)).))))..)))(((((((......)))))))((((((...))))))......))))))............... ( -32.40) >consensus GC__CUG_CA_GG__CAUCUAGAGUCAACCAACUUUAGAAGGUCAUGUGGACUUAGAGCAGCCGAAUAUCCUUGGCUGAUAUUGGAAAUGCAUAUAGC______________________ ............................((((.......(((((.....)))))....(((((((......)))))))...))))................................... (-17.08 = -16.80 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:27 2006