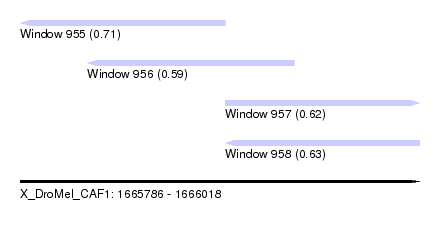
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,665,786 – 1,666,018 |
| Length | 232 |
| Max. P | 0.708871 |

| Location | 1,665,786 – 1,665,905 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.23 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -38.47 |
| Energy contribution | -38.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665786 119 - 22224390 ACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAAUCGGGCACACAUCUCAUGGCGCACACCGUGCCGCACAUAGCGUACAACAAGUUGUAGGCGCUCCAUGGCGUUACACAAC- ((((((.........))))))...(((((((((....))....)))))))...((((((.((.....(((...)))...((.(((((....))))).)))).))))))...........- ( -39.90) >DroSec_CAF1 131 119 - 1 ACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCGCACAUAGCGUACAACAAGUUGUAGGCGCUCCAUGGCGUUACACAAC- ((((((.........))))))...(((((((....((....)))))))))....(((((.((.....(((...)))...((.(((((....))))).)))).)))))............- ( -40.60) >DroSim_CAF1 988 119 - 1 ACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCGCACAUAGCGUACAACAAGUUGUAGGCGCUCCAUGGCGUUACACAAC- ((((((.........))))))...(((((((....((....)))))))))....(((((.((.....(((...)))...((.(((((....))))).)))).)))))............- ( -40.60) >DroYak_CAF1 1001 120 - 1 ACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUGCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCACACAUAACGUACAACAAGUUGUAGGCGCACCAUGGGCUUACACAACA ((((((.........))))))...(((((((((........)))))))))......((((((.....)))))).................((.((((((.(.....).)))))))).... ( -40.30) >consensus ACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCGCACAUAGCGUACAACAAGUUGUAGGCGCUCCAUGGCGUUACACAAC_ ((((((.........))))))...(((((((....((....)))))))))....(((((.((.....(((...)))...((.(((((....))))).)))).)))))............. (-38.47 = -38.72 + 0.25)

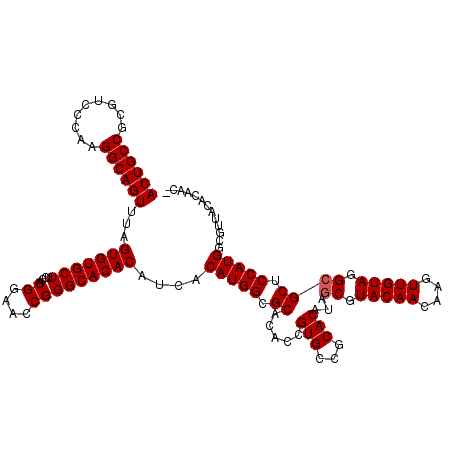
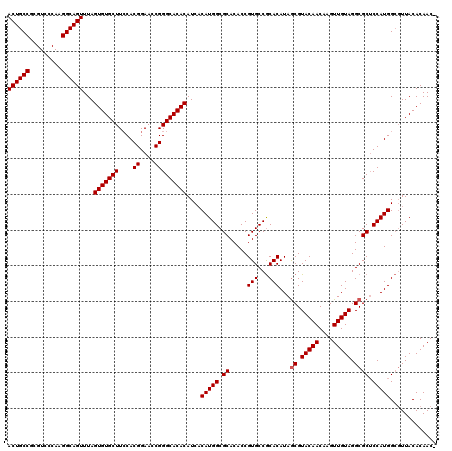
| Location | 1,665,825 – 1,665,945 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -40.79 |
| Energy contribution | -40.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665825 120 - 22224390 UCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCAACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAAUCGGGCACACAUCUCAUGGCGCACACCGUGCCGCACAUAG .(((.((.((..((((((..(((((.((...((.((..((((((((.........)))))))..)..)))).))...))))).))))))........(((((.....))))))))).))) ( -41.90) >DroSec_CAF1 170 120 - 1 UCUACGUUGCAAUGUGCCAAGGUUCAGGUUUAGUCGUGCAACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCGCACAUAG .(((.((.((..((((((..(((((.((...((.((..((((((((.........)))))))..)..)))).))...))))).))))))........(((((.....))))))))).))) ( -45.20) >DroSim_CAF1 1027 120 - 1 UCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCAACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCGCACAUAG .(((.((.((..((((((..(((((.((...((.((..((((((((.........)))))))..)..)))).))...))))).))))))........(((((.....))))))))).))) ( -44.50) >DroYak_CAF1 1041 120 - 1 UCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCAACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUGCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCACACAUAA ............((((((..(((((..((.(((.((..((((((((.........)))))))..)..)).))).)).))))).)))))).......((((((.....))))))....... ( -42.70) >consensus UCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCAACUGCCGCGUCCCAAGGCAGUUUAGUGUGCUUCCACGGAACCGGGCACACAUCACAUGGCGCACACCGUGCCGCACAUAG .....((.((..((((((..(((((.(....((.((..((((((((.........)))))))..)..))))....).))))).))))))........(((((.....))))))))).... (-40.79 = -40.85 + 0.06)

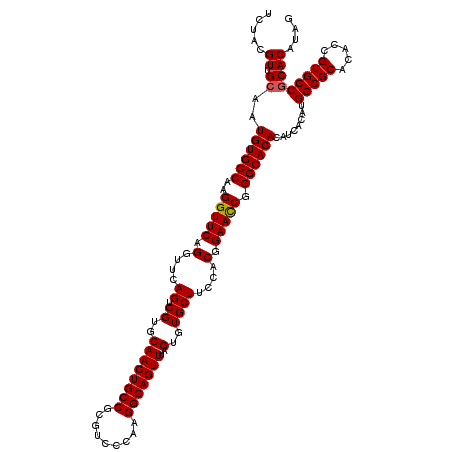
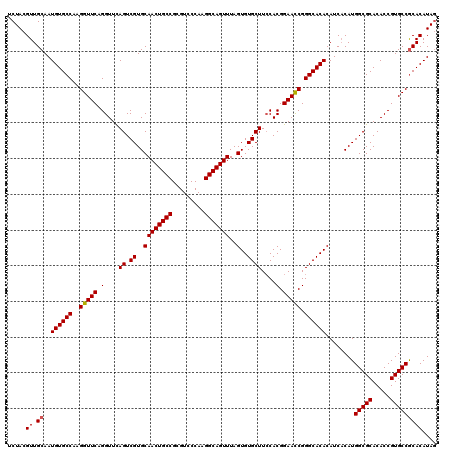
| Location | 1,665,905 – 1,666,018 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.71 |
| Mean single sequence MFE | -34.41 |
| Consensus MFE | -30.52 |
| Energy contribution | -30.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665905 113 + 22224390 UGCACGACUGAACCUGAACCUUGGCACAUUGCAACGUAGACACGUAGACACGCAGACUCGUCGCCCGAGUCGCGCGAUUGCGAUUCUCGUUGCCCAUUUCGGAUGCUAUAUAU .(((...(((((((........))......((((((.(((..(((((.(.(((.((((((.....))))))))).).)))))..)))))))))....))))).)))....... ( -35.90) >DroSec_CAF1 250 93 + 1 UGCACGACUAAACCUGAACCUUGGCACAUUGCAACGUAGACACGUAGACACGCAGACUCGUCGCCCGAGUCGCGCGAUUGCGAUUCUCGUUGC-------------------- (((.(((.............))))))....((((((.(((..(((((.(.(((.((((((.....))))))))).).)))))..)))))))))-------------------- ( -29.92) >DroSim_CAF1 1107 113 + 1 UGCACGACUGAACCUGAACCUUGGCACAUUGCAACGUAGACACGUAGACACGCAGACUCGUCGCCCGAGUCGCGCGAUUGCGAUUCUCGUUGCCCAUUUCGGAUGCUAUAUAU .(((...(((((((........))......((((((.(((..(((((.(.(((.((((((.....))))))))).).)))))..)))))))))....))))).)))....... ( -35.90) >DroYak_CAF1 1121 113 + 1 UGCACGACUGAACCUGAACCUUGGCACAUUGCAACGUAGACACGUAGACACGCAGACUCGUCGCCCGAGUCGCGCGAUUGCGAUUCUCGUUGCCCAUUUCGGAUGCUAUAUAU .(((...(((((((........))......((((((.(((..(((((.(.(((.((((((.....))))))))).).)))))..)))))))))....))))).)))....... ( -35.90) >consensus UGCACGACUGAACCUGAACCUUGGCACAUUGCAACGUAGACACGUAGACACGCAGACUCGUCGCCCGAGUCGCGCGAUUGCGAUUCUCGUUGCCCAUUUCGGAUGCUAUAUAU (((.(((.............))))))....((((((.(((..(((((.(.(((.((((((.....))))))))).).)))))..))))))))).................... (-30.52 = -30.52 + -0.00)


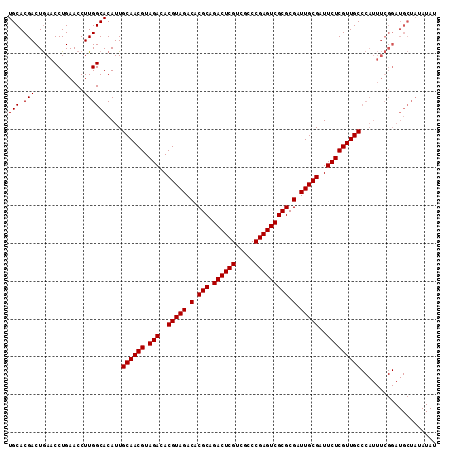
| Location | 1,665,905 – 1,666,018 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.71 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -35.24 |
| Energy contribution | -36.05 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665905 113 - 22224390 AUAUAUAGCAUCCGAAAUGGGCAACGAGAAUCGCAAUCGCGCGACUCGGGCGACGAGUCUGCGUGUCUACGUGUCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCA .......(((..(((.....(((((((((..((....(((((((((((.....)))))).)))))....))..))).))))))..((.(((........))).)).)))))). ( -39.60) >DroSec_CAF1 250 93 - 1 --------------------GCAACGAGAAUCGCAAUCGCGCGACUCGGGCGACGAGUCUGCGUGUCUACGUGUCUACGUUGCAAUGUGCCAAGGUUCAGGUUUAGUCGUGCA --------------------(((((((((..((....(((((((((((.....)))))).)))))....))..))).)))))).(((.(((........))).....)))... ( -33.20) >DroSim_CAF1 1107 113 - 1 AUAUAUAGCAUCCGAAAUGGGCAACGAGAAUCGCAAUCGCGCGACUCGGGCGACGAGUCUGCGUGUCUACGUGUCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCA .......(((..(((.....(((((((((..((....(((((((((((.....)))))).)))))....))..))).))))))..((.(((........))).)).)))))). ( -39.60) >DroYak_CAF1 1121 113 - 1 AUAUAUAGCAUCCGAAAUGGGCAACGAGAAUCGCAAUCGCGCGACUCGGGCGACGAGUCUGCGUGUCUACGUGUCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCA .......(((..(((.....(((((((((..((....(((((((((((.....)))))).)))))....))..))).))))))..((.(((........))).)).)))))). ( -39.60) >consensus AUAUAUAGCAUCCGAAAUGGGCAACGAGAAUCGCAAUCGCGCGACUCGGGCGACGAGUCUGCGUGUCUACGUGUCUACGUUGCAAUGUGCCAAGGUUCAGGUUCAGUCGUGCA .......((((.........(((((((((..((....(((((((((((.....)))))).)))))....))..))).))))))..((.(((........))).))...)))). (-35.24 = -36.05 + 0.81)

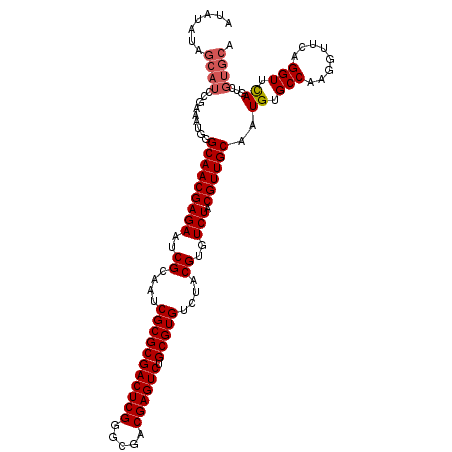
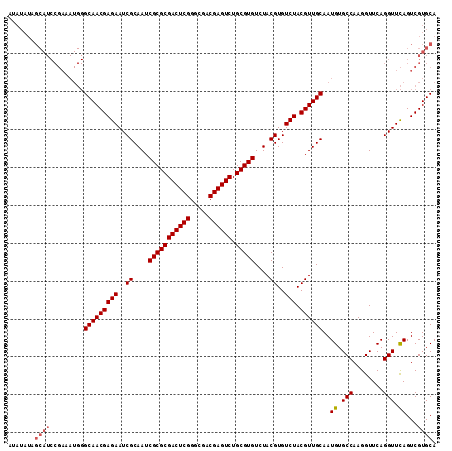
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:55 2006