| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 11,027,542 – 11,027,669 |
| Length | 127 |
| Max. P | 0.898269 |

| Location | 11,027,542 – 11,027,648 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
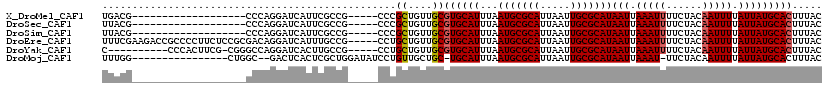
>X_DroMel_CAF1 11027542 106 - 22224390 G-----CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAU---GG------CAAAUGCUCAACAUUAAAACCAGAUUA (-----(.(((....))).)).(((((((.(((.....)))(((((((.(((((......))))).)))))))........(---((------(....)).)).)))))))......... ( -23.90) >DroSec_CAF1 19994 106 - 1 G-----CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAC---GG------CAAAUGCUCAACAUUAAAACCAGAUAC (-----((.((....))((((((...(((((((.....)))))))(((.(((((......))))).))))))))).......---))------).((((.....))))............ ( -24.00) >DroSim_CAF1 11749 106 - 1 G-----CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAC---GG------CAAAUGCUCAACAUUAAAACCAGAUUA (-----((.((....))((((((...(((((((.....)))))))(((.(((((......))))).))))))))).......---))------).((((.....))))............ ( -24.00) >DroEre_CAF1 14275 106 - 1 G-----CCUGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAU---GG------CAAAUGCUGAACAUUAAAACCAGAUUA (-----((...(((...((((((...(((((((.....)))))))(((.(((((......))))).)))))))))...))).---))------).....(((...........))).... ( -24.10) >DroYak_CAF1 15737 106 - 1 G-----CCUGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAU---GG------CAAAUGCUGAACAUUAAAACCAGAUUA (-----((...(((...((((((...(((((((.....)))))))(((.(((((......))))).)))))))))...))).---))------).....(((...........))).... ( -24.10) >DroMoj_CAF1 18357 118 - 1 GGAUAUCCUGUUGCUGC-UGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAU-UUCUACAAUUUUAUUAUGCACUUUACAGCCUGACGCGCUCCGAUGCUCAGCAUUAAAAUCAGAUUA ((....))...(((((.-.(((((....((((.........(((((((.((((-(.....))))).)))))))......(.....)..))))...))))).))))).............. ( -27.50) >consensus G_____CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUACAU___GG______CAAAUGCUCAACAUUAAAACCAGAUUA ...........(((((...((((((.....(((.....)))(((((((.(((((......))))).))))))).....................)))))).))))).............. (-20.15 = -20.40 + 0.25)

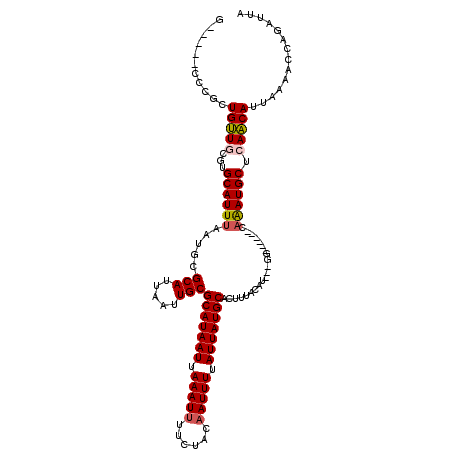
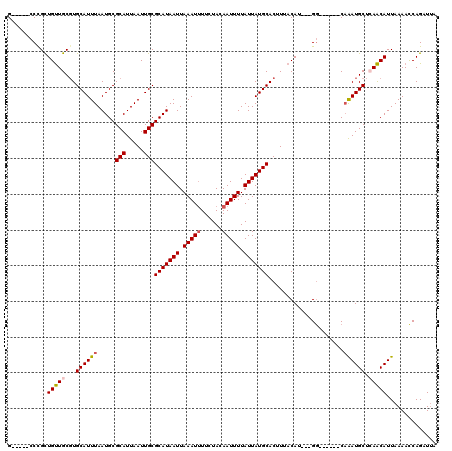
| Location | 11,027,573 – 11,027,669 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11027573 96 + 22224390 GUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGCAACAGCGGG-----CGGCGAAUGAUCCUGGG-------------------CGUCA .....(((((((((.(((((......))))).))))))))).......((((((...)))).(((....))).)-----)((((..(......)..-------------------)))). ( -26.10) >DroSec_CAF1 20025 96 + 1 GUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGCAACAGCGGG-----CGGCGAAUGAUCCUGGG-------------------CGUAA .....(((((((((.(((((......))))).)))))))))......(((((.......((.(((....))).)-----)((........))...)-------------------)))). ( -26.90) >DroSim_CAF1 11780 96 + 1 GUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGCAACAGCGGG-----CGGCGAAUGAUCCUGGG-------------------CGUAA .....(((((((((.(((((......))))).)))))))))......(((((.......((.(((....))).)-----)((........))...)-------------------)))). ( -26.90) >DroEre_CAF1 14306 115 + 1 GUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGCAACAGCAGG-----CGGCAAAUGAUCCUGUCGCGGAGAAGGGGCGGUCUUCGAAA .....(((((((((.(((((......))))).)))(((((((.....)))))))...))))))((....))..(-----(((((........))))))(((((........))))).... ( -32.00) >DroYak_CAF1 15768 104 + 1 GUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGCAACAGCAGG-----CGGCAAGUGAUCCUGGCCCG-CGAAGUGGG----------G .....(((((((((.(((((......))))).)))))))))......(((((((...)))).(((........)-----)))))...........((((-(...)))))----------. ( -29.10) >DroMoj_CAF1 18397 100 + 1 GUAAAGUGCAUAAUAAAAUUGUAGAA-AUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCA-GCAGCAACAGGAUAUCCAGCGAGUGAGUC--GCCAG----------------CCAAA .....(((((((((.(((((.....)-)))).)))))))))......(((((((...)))).-)))((....((....)).((((.....))--))..)----------------).... ( -25.90) >consensus GUAAAGUGCAUAAUAAAAUUGUAGAAAAUUUAAUUAUGCGCAAUUAAUGCGCAUUAAAUGCACGCAACAGCAGG_____CGGCGAAUGAUCCUGGC___________________CGUAA .....(((((((((.(((((......))))).)))))))))......(((((((...))))..((....))..........))).................................... (-18.91 = -19.02 + 0.11)
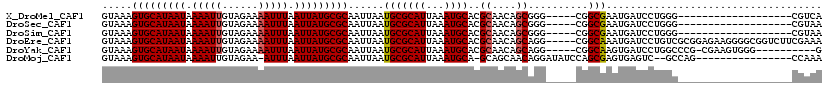
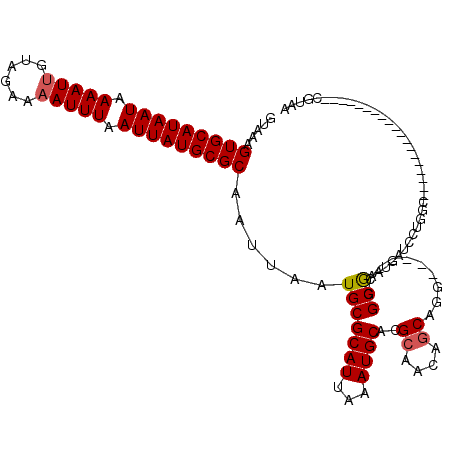
| Location | 11,027,573 – 11,027,669 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 11027573 96 - 22224390 UGACG-------------------CCCAGGAUCAUUCGCCG-----CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUAC ....(-------------------(...((........))(-----(.(((....))).)).......))(((.....)))(((((((.(((((......))))).)))))))....... ( -21.20) >DroSec_CAF1 20025 96 - 1 UUACG-------------------CCCAGGAUCAUUCGCCG-----CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUAC ....(-------------------(...((........))(-----(.(((....))).)).......))(((.....)))(((((((.(((((......))))).)))))))....... ( -21.20) >DroSim_CAF1 11780 96 - 1 UUACG-------------------CCCAGGAUCAUUCGCCG-----CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUAC ....(-------------------(...((........))(-----(.(((....))).)).......))(((.....)))(((((((.(((((......))))).)))))))....... ( -21.20) >DroEre_CAF1 14306 115 - 1 UUUCGAAGACCGCCCCUUCUCCGCGACAGGAUCAUUUGCCG-----CCUGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUAC ....((((.......))))...(((((((.(..........-----..).)))))))((((((...(((((((.....)))))))(((.(((((......))))).)))))))))..... ( -28.80) >DroYak_CAF1 15768 104 - 1 C----------CCCACUUCG-CGGGCCAGGAUCACUUGCCG-----CCUGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUAC .----------........(-(((((((((....))))..)-----)))))......((((((...(((((((.....)))))))(((.(((((......))))).)))))))))..... ( -29.80) >DroMoj_CAF1 18397 100 - 1 UUUGG----------------CUGGC--GACUCACUCGCUGGAUAUCCUGUUGCUGC-UGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAU-UUCUACAAUUUUAUUAUGCACUUUAC ...((----------------(.(((--(((.........((....)).))))))))-)(((.(((((....))))).)))(((((((.((((-(.....))))).)))))))....... ( -23.80) >consensus UUACG___________________CCCAGGAUCAUUCGCCG_____CCCGCUGUUGCGUGCAUUUAAUGCGCAUUAAUUGCGCAUAAUUAAAUUUUCUACAAUUUUAUUAUGCACUUUAC .................................................((....))((((((...(((((((.....)))))))(((.(((((......))))).)))))))))..... (-17.16 = -17.35 + 0.19)
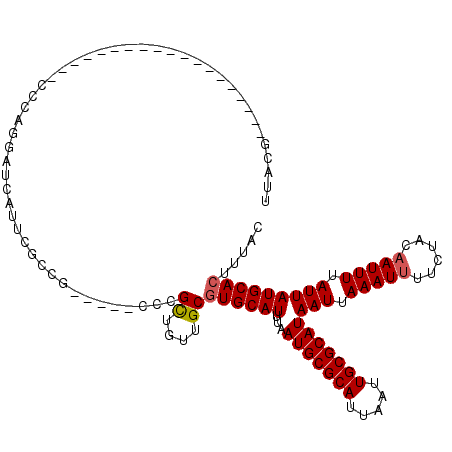
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:09 2006