| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,436,452 – 9,436,571 |
| Length | 119 |
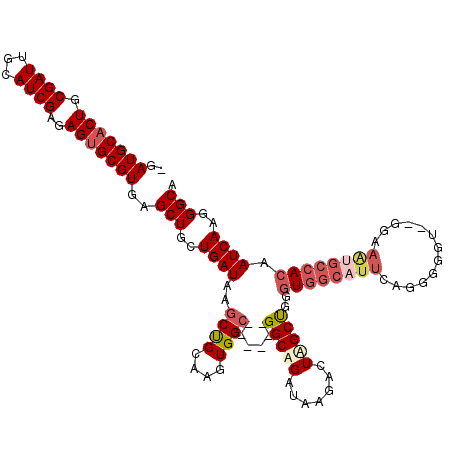
| Max. P | 0.807600 |

| Location | 9,436,452 – 9,436,571 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -16.73 |
| Energy contribution | -18.29 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9436452 119 + 22224390 UGCCCUUGAUUGUGGCAUUUCUGAACCAUCUGAAUGCCACACAGCUAGUCUUAUCUGCCUUAGCUGCCACUUGCAGCUAAUCAGCAGCUUACGCACUCUCGAUGCAAUCGCAGUGCAUU- ..........((((((((((..((....)).)))))))))).............((((.((((((((.....))))))))...)))).....(((((..((((...)))).)))))...- ( -37.40) >DroSec_CAF1 18056 118 + 1 UGCCCUUGAUUGUGGCACUUCC--ACCCCCUGAAUGCCACCCGGCUAGUCUUAUCUGCCUUAGCUGCCACUUGCAGCUUAUCAGCAGCUCACGCACUCUCGAUGCAAUCGCAGUGCAUCG .((..(((((.((((((.(((.--.......)))))))))..(((.((......)))))..((((((.....)))))).)))))..))....(((((..((((...)))).))))).... ( -35.00) >DroEre_CAF1 18431 97 + 1 UGCCCUUGAUUGUGGCAUUUCC----------------AUCCAGCCAUUCUUAUCAGCC------UCCACUUGCAGCUUAUCAGCAGCUCACGCACUCUCGAUGCAAUCGCAGUGCAUC- .....(((((.(((((......----------------.....)))))....)))))..------.......((.(((....))).))....(((((..((((...)))).)))))...- ( -20.10) >DroYak_CAF1 18981 111 + 1 UGCCCUUGAUUGUGGCAUUUCC--AGCCCCUGAAAGCCACCCAGCUACUCUUAUCAGCC------GCCACUUGCGGCUUAUCAGCAGCUCACGCCCUCUCGAUGCAAUCGCAGUGCAUC- (((.((((((((((((.((((.--.......)))))))....((((.((..((..((((------((.....))))))))..)).))))..............))))))).)).)))..- ( -31.10) >consensus UGCCCUUGAUUGUGGCAUUUCC__ACCCCCUGAAUGCCACCCAGCUAGUCUUAUCAGCC______GCCACUUGCAGCUUAUCAGCAGCUCACGCACUCUCGAUGCAAUCGCAGUGCAUC_ (((.(((((((((((((..............((..((......))...))..............))))))..((((((....))).((....))........)))))))).)).)))... (-16.73 = -18.29 + 1.56)

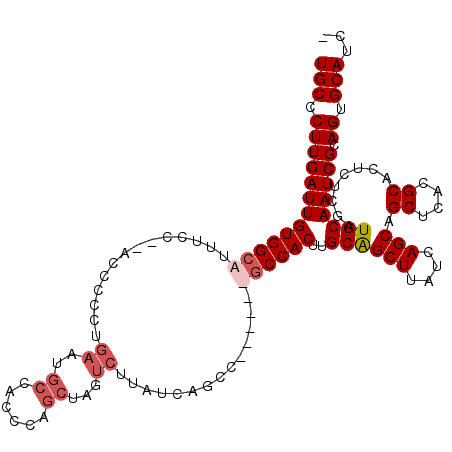
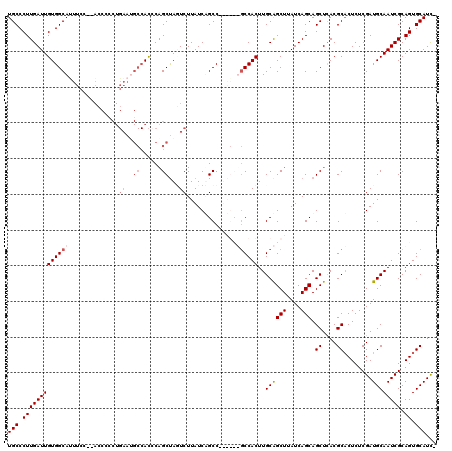
| Location | 9,436,452 – 9,436,571 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -23.11 |
| Energy contribution | -25.55 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9436452 119 - 22224390 -AAUGCACUGCGAUUGCAUCGAGAGUGCGUAAGCUGCUGAUUAGCUGCAAGUGGCAGCUAAGGCAGAUAAGACUAGCUGUGUGGCAUUCAGAUGGUUCAGAAAUGCCACAAUCAAGGGCA -.(((((((.((((...))))..)))))))...(((((..((((((((.....))))))))))))).........(((.((((((((((.((....)).).)))))))))......))). ( -44.50) >DroSec_CAF1 18056 118 - 1 CGAUGCACUGCGAUUGCAUCGAGAGUGCGUGAGCUGCUGAUAAGCUGCAAGUGGCAGCUAAGGCAGAUAAGACUAGCCGGGUGGCAUUCAGGGGGU--GGAAGUGCCACAAUCAAGGGCA ..(((((((.((((...))))..)))))))..(((..((((.((((((.....))))))..(((((......)).)))..(((((((((.......--.).)))))))).))))..))). ( -43.10) >DroEre_CAF1 18431 97 - 1 -GAUGCACUGCGAUUGCAUCGAGAGUGCGUGAGCUGCUGAUAAGCUGCAAGUGGA------GGCUGAUAAGAAUGGCUGGAU----------------GGAAAUGCCACAAUCAAGGGCA -((((((.......))))))....((.(.(((((.(((....))).))..((((.------((((.........))))..((----------------....)).))))..))).).)). ( -25.70) >DroYak_CAF1 18981 111 - 1 -GAUGCACUGCGAUUGCAUCGAGAGGGCGUGAGCUGCUGAUAAGCCGCAAGUGGC------GGCUGAUAAGAGUAGCUGGGUGGCUUUCAGGGGCU--GGAAAUGCCACAAUCAAGGGCA -((((((.......))))))((...(((((.((((.((((.(((((((.(((.((------..((....)).)).)))..))))))))))).))))--....)))))....))....... ( -43.80) >consensus _GAUGCACUGCGAUUGCAUCGAGAGUGCGUGAGCUGCUGAUAAGCUGCAAGUGGC______GGCAGAUAAGACUAGCUGGGUGGCAUUCAGGGGGU__GGAAAUGCCACAAUCAAGGGCA ..(((((((.((((...))))..)))))))..(((..((((..((((....))))......(((((.......)))))..((((((((.............)))))))).))))..))). (-23.11 = -25.55 + 2.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:27 2006