| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,049,024 – 8,049,129 |
| Length | 105 |
| Max. P | 0.650809 |

| Location | 8,049,024 – 8,049,129 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 51.59 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -5.55 |
| Energy contribution | -3.73 |
| Covariance contribution | -1.83 |
| Combinations/Pair | 2.20 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650809 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
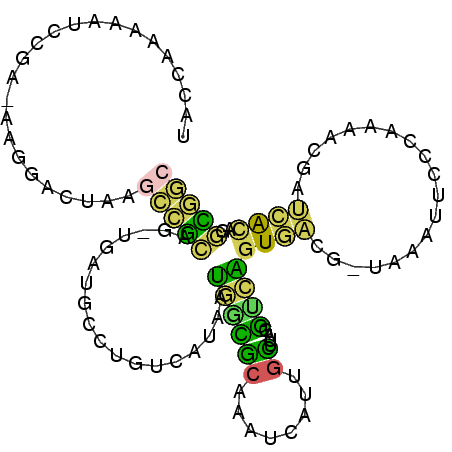
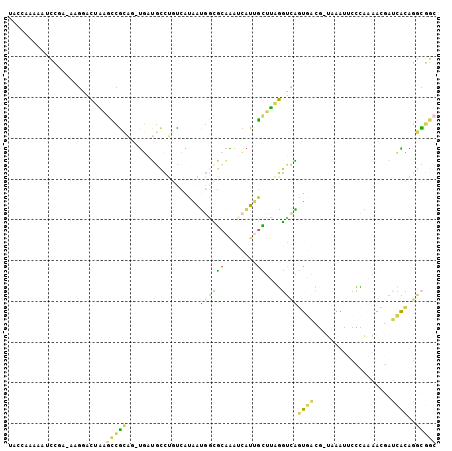
>X_DroMel_CAF1 8049024 105 + 22224390 UACCAAAAAUCCAAAAAGGACUAAGCCGCACUUGCUGCCUGUCAUAAUGACGCAAAUCAUUGCUUAGGUCAGUGACGGUGAAUACCCAAAACGAUCACAGGCGGC .........(((.....)))....(((((..(..(((((((((.....)))((((....))))..))).)))..)..((((.............))))..))))) ( -27.72) >DroVir_CAF1 18568 102 + 1 ACACUCAUCUUUGC-GAUGUAUGUAUGUUCG-UAGAUGUGAGCAUGUAUGUGCUGGAUAAUGUACAUGCGUGUACAU-AUGAAUGAAUAUGCAUGUGUACAAAAA ((((.((((.....-)))).(((((((((((-(..(((((.((((((((((((........)))))))))))).)))-))..))))))))))))))))....... ( -40.70) >DroSec_CAF1 24680 102 + 1 UACCAAAAAACCAA-GAGGACUAAGCCGCAG-UGAUGCCUCUCAUAAUGGCGCAAAUCAUAGCUUAGGUCAGUGACG-UAAAUUCCCAAAACGAUCACAGGCGGC ..........((..-..)).....(((((..-(((((((.........)))((........))....))))((((((-(...........))).))))..))))) ( -23.30) >DroEre_CAF1 19206 101 + 1 UACCAAA-ACCCGA-AAGCACUAAGCCGCAG-UGAUGCCUGUCAUAAUGGUGCAAAUCAUUGCUUGGGUCAGUGACG-UAAAUUCCCAAAAGGAUCACAGGCGGC .......-...(..-..)......(((((.(-((((.(((....(((((((....))))))).(((((.........-......))))).))))))))..))))) ( -27.76) >DroWil_CAF1 23611 102 + 1 UACAUAGAUUAAUC-UAAGGAAUAGAUGCAC-CAAGAGACAUAGAGAGAUGCCAGAUCUUUUGGAAUUUCCAUACAU-ACACAGAAGCAUGUAAAAGGAUAUACA ...........(((-((.....)))))...(-((((((((((......))).....))))))))....(((.(((((-.(......).)))))...)))...... ( -14.60) >DroYak_CAF1 24807 102 + 1 UAACAAAUAACCGA-AAGGACUAAGCCGCAG-UGAUGCCUGCCAUAAUGGCGCAAAUCAUUGCUUAGGUCAGUGACG-UAAACUCCCAAAACGAUCAUAGGCGGC ..........((..-..)).....(((((((-((((...((((.....))))...))))))))....(((.((((((-(...........))).)))).)))))) ( -32.80) >consensus UACCAAAAAUCCGA_AAGGACUAAGCCGCAG_UGAUGCCUGUCAUAAUGGCGCAAAUCAUUGCUUAGGUCAGUGACG_UAAAUUCCCAAAACGAUCACAGGCGGC ........................(((((..................((((((........))....))))((((...................))))..))))) ( -5.55 = -3.73 + -1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:56 2006