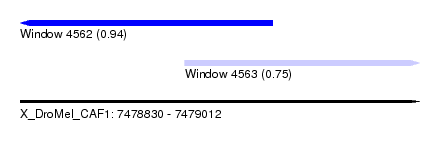
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,478,830 – 7,479,012 |
| Length | 182 |
| Max. P | 0.938021 |

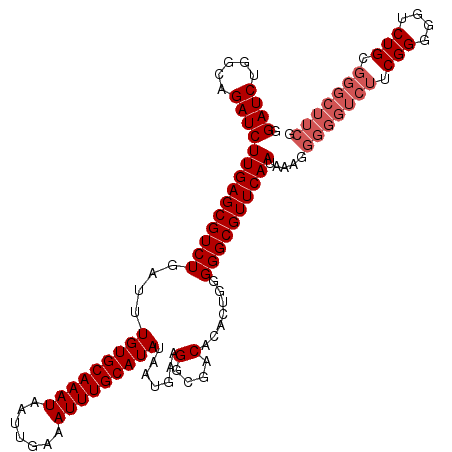
| Location | 7,478,830 – 7,478,945 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.28 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -28.99 |
| Energy contribution | -30.92 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7478830 115 - 22224390 GGAUCUGGCAGAUCUUGAGCGUCUGAUUUGUGCAAAUAAUUGAAAUUUGCAUAUAAUGAAGGCGACACACUGGGGGCGUUCAAUAAAGGGGGUCUUCGGGGGACUGCGGGCUUCG (((.((.((((.((((..((((((....(((((((((.......)))))))))......))))).)...((((((((.(((......))).)))))))))))))))).)).))). ( -40.40) >DroSec_CAF1 11971 115 - 1 GGAUCUGGCAGAUCUUGAGCGUCUGAUUUGUGCAAAUAAUUGAAAUUUGCAUAUAAUGAAGGCGACACACUGGGGGCGUUCAAUAAAGGGGGUCUUCGGGGGUCUGCGGGCUUCG (((.((.((((((((...((((((....(((((((((.......)))))))))......))))).)...((((((((.(((......))).)))))))))))))))).)).))). ( -43.80) >DroSim_CAF1 11998 115 - 1 GGAUCUGGCAGAUCUUGAGCGUCUGAUUUGUGCAAAUAAUUGAAAUUUGCAUAUAAUGAAGGCGACACACUGGGGGCGUUCAAUAAAGGGGGUCUUCGGGGGUCUGCGGGCUUCG (((.((.((((((((...((((((....(((((((((.......)))))))))......))))).)...((((((((.(((......))).)))))))))))))))).)).))). ( -43.80) >DroEre_CAF1 12808 96 - 1 GGAUCUGGCAGAUCUUGAGCGUCUGAUUUGUGCAAAUAAUUGAAAUUUGCAUAUAAUGAAGGCGACACACUGGGGGCGUUCAAUAAAAGCGGGUCU------------------- (((((((.(.....((((((((((....(((((((((.......))))))))).......(....).......)))))))))).....))))))))------------------- ( -27.80) >consensus GGAUCUGGCAGAUCUUGAGCGUCUGAUUUGUGCAAAUAAUUGAAAUUUGCAUAUAAUGAAGGCGACACACUGGGGGCGUUCAAUAAAGGGGGUCUUCGGGGGUCUGCGGGCUUCG .((((.....))))((((((((((....(((((((((.......))))))))).......(....).......)))))))))).....(((((((.(((....))).))))))). (-28.99 = -30.92 + 1.94)

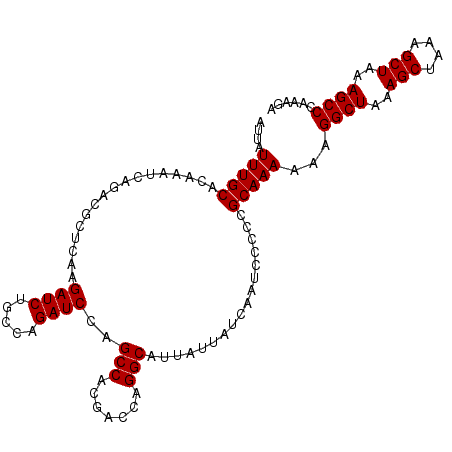
| Location | 7,478,905 – 7,479,012 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 99.53 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7478905 107 + 22224390 AUUAUUUGCACAAAUCAGACGCUCAAGAUCUGCCAGAUCCAGCCACGACCAGGCAUUAUUAUCAAUCCCCCGCAAAAAAGGCUAAAGCUAAAGCUAAAGCCCAAAGA ....(((((.................((((.....))))..(((.......))).................)))))...((((..(((....)))..))))...... ( -21.10) >DroSec_CAF1 12046 107 + 1 AUUAUUUGCACAAAUCAGACGCUCAAGAUCUGCCAGAUCCAGCCACGACCAGGCAUUAUUAUCAAUCCCCCGCAAAAAAGGCUAAAGCUUAAGCUAAAGCCCAAAGA ....(((((.................((((.....))))..(((.......))).................)))))...((((..(((....)))..))))...... ( -21.10) >DroSim_CAF1 12073 107 + 1 AUUAUUUGCACAAAUCAGACGCUCAAGAUCUGCCAGAUCCAGCCACGACCAGGCAUUAUUAUCAAUCCCCCGCAAAAAAGGCUAAAGCUAAAGCUAAAGCCCAAAGA ....(((((.................((((.....))))..(((.......))).................)))))...((((..(((....)))..))))...... ( -21.10) >DroEre_CAF1 12864 107 + 1 AUUAUUUGCACAAAUCAGACGCUCAAGAUCUGCCAGAUCCAGCCACGACCAGGCAUUAUUAUCAAUCCCCCGCAAAAAAGGCUAAAGCUAAAGCUAAAGCCCAAAGA ....(((((.................((((.....))))..(((.......))).................)))))...((((..(((....)))..))))...... ( -21.10) >consensus AUUAUUUGCACAAAUCAGACGCUCAAGAUCUGCCAGAUCCAGCCACGACCAGGCAUUAUUAUCAAUCCCCCGCAAAAAAGGCUAAAGCUAAAGCUAAAGCCCAAAGA ....(((((.................((((.....))))..(((.......))).................)))))...((((..(((....)))..))))...... (-21.10 = -21.10 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:21 2006