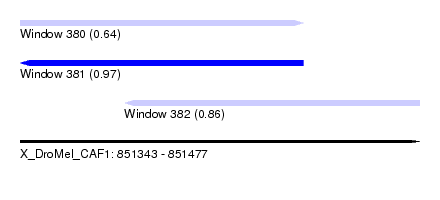
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 851,343 – 851,477 |
| Length | 134 |
| Max. P | 0.972564 |

| Location | 851,343 – 851,438 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -12.83 |
| Energy contribution | -13.63 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
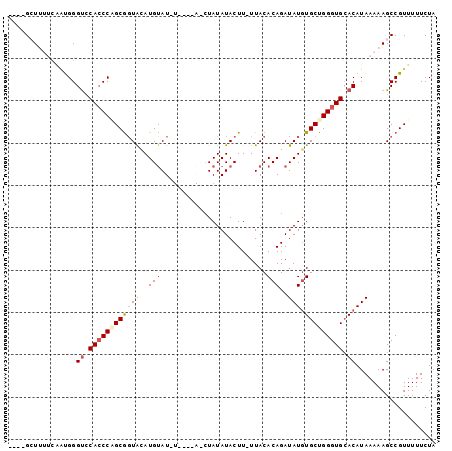
>X_DroMel_CAF1 851343 95 + 22224390 ----GCUUUUC-AUGGGUCCAACCGGCGGUGCAUGUAUCUAUGUAUGUAUAUACUU-UUACACAGAUAUUUGCUGGGUGAACAUAAAAAGCCGUUUUUCUA ----((((((.-(((..((...(((((((.....((((((.((((.((....))..-.)))).)))))))))))))..)).))).)))))).......... ( -24.90) >DroSec_CAF1 54121 93 + 1 ----GCUUUUCAUCGGGUCCACCCAGCAGUACAUGUAUAU----ACCUAUAUACUUUUUACACAGAUAUGUGCUGGGUGCACAUAAAAAGCCGUUUUUCUA ----((((((......((.((((((((.(((...((((((----....))))))....)))(((....))))))))))).))...)))))).......... ( -27.10) >DroSim_CAF1 50323 92 + 1 ----GCUUUUGCAUGGGUCCACCCAGCGGUACAUGUAUAU----ACCUAUAUACUU-UUACACAGAUAUGUGCUGGGUGCACAUAAAAAGCCGUUUUUCUA ----((((((..(((....((((((((.(((...((((((----....))))))..-.)))(((....)))))))))))..))).)))))).......... ( -29.20) >DroEre_CAF1 54072 91 + 1 CAAGGAUAG-CAGUGGGCCCACCCUGCGCUAUAUGUAU--------CUAUAUACUU-UUACACAGAUAUGUGCUGGGUGCACAUAAAAAGCCGUUUUUCUA ..((((..(-(((.((.....))))))(((...((((.--------..........-.))))....(((((((.....)))))))...))).....)))). ( -22.20) >DroYak_CAF1 51858 87 + 1 ----GAUAG-CAGCGGGUUCACCCUGCGGUACAUGCAU--------CUAUAUACUU-UUACACAGAUAUGUGCUGGGUGCACAUAAAAAGCCGUUUUUCUA ----....(-(((.((.....))))))(((.......(--------((........-......)))(((((((.....)))))))....)))......... ( -20.34) >consensus ____GCUUUUCAAUGGGUCCACCCAGCGGUACAUGUAU_U____A_CUAUAUACUU_UUACACAGAUAUGUGCUGGGUGCACAUAAAAAGCCGUUUUUCUA ................((.((((((((((((..(((.........................)))..))).))))))))).))................... (-12.83 = -13.63 + 0.80)


| Location | 851,343 – 851,438 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -14.50 |
| Energy contribution | -15.62 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
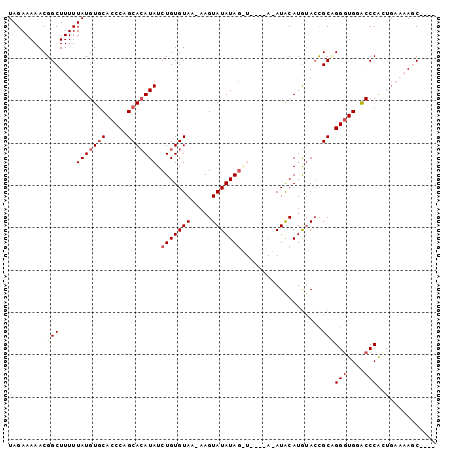
>X_DroMel_CAF1 851343 95 - 22224390 UAGAAAAACGGCUUUUUAUGUUCACCCAGCAAAUAUCUGUGUAA-AAGUAUAUACAUACAUAGAUACAUGCACCGCCGGUUGGACCCAU-GAAAAGC---- ..........((((((((((.....((((((..((((((((((.-..((....)).))))))))))..)))(((...))))))...)))-)))))))---- ( -24.90) >DroSec_CAF1 54121 93 - 1 UAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAAAAAGUAUAUAGGU----AUAUACAUGUACUGCUGGGUGGACCCGAUGAAAAGC---- ..........(((((((((((.(((((((((.(((((((((((.....)))))))))----)).((....)).))))))))).))...)))))))))---- ( -33.40) >DroSim_CAF1 50323 92 - 1 UAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA-AAGUAUAUAGGU----AUAUACAUGUACCGCUGGGUGGACCCAUGCAAAAGC---- ..........(((((((((((.((((((((.......((((((.-...))))))(((----(((....)))))))))))))).))..))).))))))---- ( -30.60) >DroEre_CAF1 54072 91 - 1 UAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA-AAGUAUAUAG--------AUACAUAUAGCGCAGGGUGGGCCCACUG-CUAUCCUUG .........((.......(((((.....)))))((((((((((.-...)))))))--------)))...((((((..(((....)))..))-))))))... ( -24.40) >DroYak_CAF1 51858 87 - 1 UAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA-AAGUAUAUAG--------AUGCAUGUACCGCAGGGUGAACCCGCUG-CUAUC---- ........((((....(((((((.....)))))))((((((((.-...)))))))--------)(((.......)))(((....)))))))-.....---- ( -26.00) >consensus UAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA_AAGUAUAUAG_U____A_AUACAUGUACCGCAGGGUGGACCCACUGAAAAGC____ .........(((....(((((((.....))))))).(((((((.....)))))))...................)))(((....))).............. (-14.50 = -15.62 + 1.12)

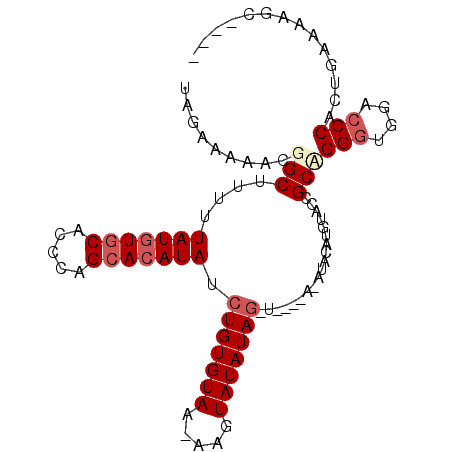
| Location | 851,378 – 851,477 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 851378 99 - 22224390 CACAACGCAACUGUAGCACACACCGAAAUGCAGCUGACAUAGAAAAACGGCUUUUUAUGUUCACCCAGCAAAUAUCUGUGUAA-AAGUAUAUACAUACAU ....(((((..(((.(((..........))).(((((((((((((......))))))))).....))))..)))..)))))..-..((((....)))).. ( -17.40) >DroSec_CAF1 54157 96 - 1 CUCAUCACAACUGUAGCACACACCGAAAUGCAGCUGACAUAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAAAAAGUAUAUAGGU---- ..........((((((((..........))).(((.(((((((............(((((((.....))))))))))))))....))).)))))..---- ( -20.40) >DroSim_CAF1 50359 95 - 1 CUCAUCACAACUGUAGCACACACCGAAAUGCAGCUGACAUAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA-AAGUAUAUAGGU---- ..........((((((((..........))).(((.(((((((............(((((((.....))))))))))))))..-.))).)))))..---- ( -21.00) >DroEre_CAF1 54109 93 - 1 CUCAUCGCAACUGUAGCACACACCGAAAUGCAGCUGACAUAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA-AAGUAUAUAG------ ..........((((((((..........))).(((.(((((((............(((((((.....))))))))))))))..-.))).)))))------ ( -19.70) >DroYak_CAF1 51891 93 - 1 CUCAUCGCAACUGUAGCACACACCGAAAUGCAGCUGACAUAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA-AAGUAUAUAG------ ..........((((((((..........))).(((.(((((((............(((((((.....))))))))))))))..-.))).)))))------ ( -19.70) >consensus CUCAUCGCAACUGUAGCACACACCGAAAUGCAGCUGACAUAGAAAAACGGCUUUUUAUGUGCACCCAGCACAUAUCUGUGUAA_AAGUAUAUAG_U____ ..........(((((..((((((.((.(((..(((((((((((((......))))))))).....)))).))).)).)))).....)).)))))...... (-16.80 = -17.20 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:02 2006