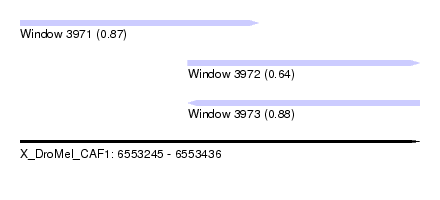
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,553,245 – 6,553,436 |
| Length | 191 |
| Max. P | 0.882260 |

| Location | 6,553,245 – 6,553,359 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -24.85 |
| Energy contribution | -24.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6553245 114 + 22224390 GGUUUAGCACUCGUUGUACAAAUAUGUACUGCAAAUUGCUUUGGUCUUUUUACCAGUUCCAGUUUCCAGGUGCUAUUCCCCUCUCAUGUUCGCUCUCGC------UCUCUGUGCAUCAUU ((..(((((((.((.(((((....))))).))((((((..(((((......)))))...))))))...)))))))..))..................((------.......))...... ( -26.30) >DroSim_CAF1 7946 114 + 1 GGUUUAGCACUCGUUGUACAAAUAUGUAUUGCAAAUUGCUUUGGUCUUUUUACCAGUUCCAGUUUCCAGGUGCUAUUCCCCUCUCAUGUUCGCUCUCGC------UCUCUGUGCAUCAUU ((..(((((((.((.(((((....))))).))((((((..(((((......)))))...))))))...)))))))..))..................((------.......))...... ( -24.90) >DroYak_CAF1 8358 120 + 1 GGUUUAGCACUCGUUGUACAAAUAUGUAUUGCAAAUUGCUUUGGUCUUUUUACCAGUUCCAGUUUCCAGGUGCUAUUCCCCUCUCUUGUUCGCUCUCGCUCUUGCUCUCUGUGCAUCAUU ((..(((((((.((.(((((....))))).))((((((..(((((......)))))...))))))...)))))))..))............((....))...(((.......)))..... ( -26.00) >consensus GGUUUAGCACUCGUUGUACAAAUAUGUAUUGCAAAUUGCUUUGGUCUUUUUACCAGUUCCAGUUUCCAGGUGCUAUUCCCCUCUCAUGUUCGCUCUCGC______UCUCUGUGCAUCAUU ((..(((((((.((.(((((....))))).))((((((..(((((......)))))...))))))...)))))))..))............((.(...............).))...... (-24.85 = -24.63 + -0.22)

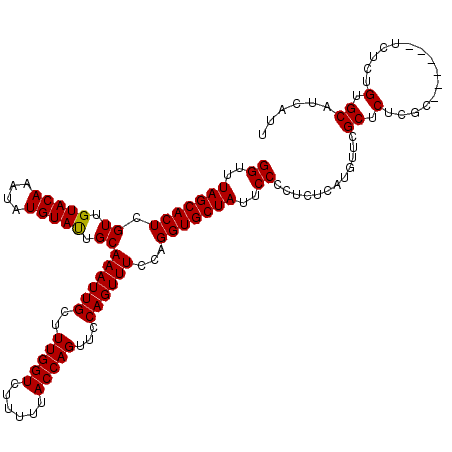
| Location | 6,553,325 – 6,553,436 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -28.89 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6553325 111 + 22224390 CUCUCAUGUUCGCUCUCGC------UCUCUGUGCAUCAUUAUGCGAGAGCUCAUUCUUGUUCUCUUUCCCCAAUGCUGAUGGGGGG---AAAGAGAGAUCGCGCUGAAGAUCAAAACAGA ......((((.((((((((------....((.....))....))))))))..........(((((((((((.((....))..))))---)))))))((((........))))..)))).. ( -39.10) >DroSim_CAF1 8026 114 + 1 CUCUCAUGUUCGCUCUCGC------UCUCUGUGCAUCAUUAUGCGAGAGCUCAUUCUUGUUCUCUUUCCCCAAUGCAGAUGGGGGGGUUAAAGAGAGAUCGCGCUGAAGAUCAAAACAGA .((((......((((((((------....((.....))....))))))))..............(..(((((.......)))))..).....))))((((........))))........ ( -34.50) >DroYak_CAF1 8438 119 + 1 CUCUCUUGUUCGCUCUCGCUCUUGCUCUCUGUGCAUCAUUAUGCGAGAGCUUAUUCUUGUUCUCUUUCCCCAAUGCAGAUGGAGGA-AAAAAGAGAGAUCGCGCAGAAGAUCAAAACAGA .((((((....((((((((...(((.......))).......))))))))..............((((((((.......))).)))-)).))))))((((........))))........ ( -35.30) >consensus CUCUCAUGUUCGCUCUCGC______UCUCUGUGCAUCAUUAUGCGAGAGCUCAUUCUUGUUCUCUUUCCCCAAUGCAGAUGGGGGG___AAAGAGAGAUCGCGCUGAAGAUCAAAACAGA ......((((.((((((((.......................))))))))..........((((((((((((.......))))))).....)))))((((........))))..)))).. (-28.89 = -29.00 + 0.11)

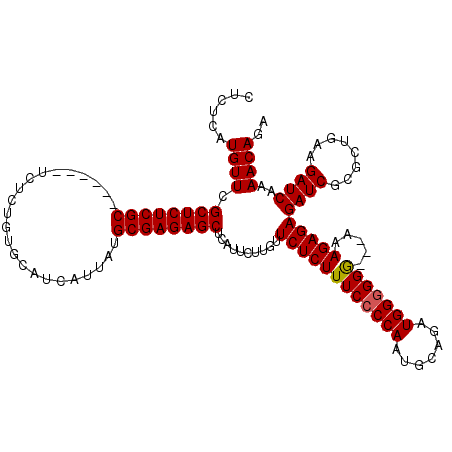
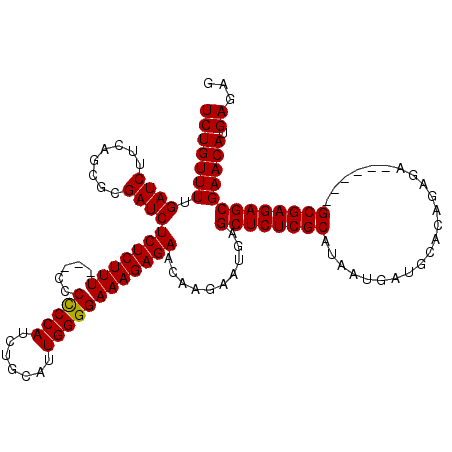
| Location | 6,553,325 – 6,553,436 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6553325 111 - 22224390 UCUGUUUUGAUCUUCAGCGCGAUCUCUCUUU---CCCCCCAUCAGCAUUGGGGAAAGAGAACAAGAAUGAGCUCUCGCAUAAUGAUGCACAGAGA------GCGAGAGCGAACAUGAGAG ..........(((.((.(((..(((((((((---((((...........)))))))))))..........((((((((((....))))...))))------))))..)))....))))). ( -39.70) >DroSim_CAF1 8026 114 - 1 UCUGUUUUGAUCUUCAGCGCGAUCUCUCUUUAACCCCCCCAUCUGCAUUGGGGAAAGAGAACAAGAAUGAGCUCUCGCAUAAUGAUGCACAGAGA------GCGAGAGCGAACAUGAGAG ..........(((.((.(((..(((((((((.....(((((.......))))))))))))..........((((((((((....))))...))))------))))..)))....))))). ( -35.90) >DroYak_CAF1 8438 119 - 1 UCUGUUUUGAUCUUCUGCGCGAUCUCUCUUUUU-UCCUCCAUCUGCAUUGGGGAAAGAGAACAAGAAUAAGCUCUCGCAUAAUGAUGCACAGAGAGCAAGAGCGAGAGCGAACAAGAGAG .((.(((((.((((((.(((....((((((...-(((.(((.......))))))))))))..........((((((((((....))))...))))))....))))))).)).))))).)) ( -38.50) >consensus UCUGUUUUGAUCUUCAGCGCGAUCUCUCUUU___CCCCCCAUCUGCAUUGGGGAAAGAGAACAAGAAUGAGCUCUCGCAUAAUGAUGCACAGAGA______GCGAGAGCGAACAUGAGAG (((((((.((((........))))(((((((.....(((((.......))))))))))))..........((((((((.......................))))))))))))).))... (-32.22 = -32.00 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:36 2006