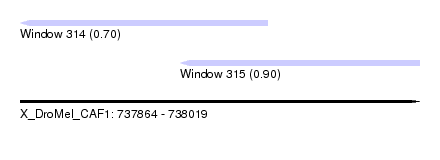
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 737,864 – 738,019 |
| Length | 155 |
| Max. P | 0.896866 |

| Location | 737,864 – 737,960 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -19.77 |
| Energy contribution | -19.05 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
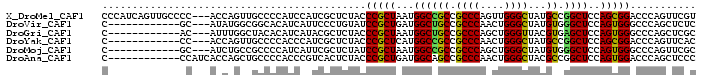
>X_DroMel_CAF1 737864 96 - 22224390 CCCAUCAGUUGCCCC---ACCAGUUGCCCCAUCCAUCGCUCUACCCGCUAAUGGCCGCCGCCCAGUUGGGCUAUGCCGGCUCCAGCGGACCCAGUUCGU .....((..((....---..))..))...........(((....(((((...((((((.((((....))))...).)))))..)))))....))).... ( -23.40) >DroVir_CAF1 86653 84 - 1 C------------GC---AUAUGGCGGCACAUCAUUCCCUGUAUCCGCUGAUGGCUGCCGCCCAACUGGGCUAUGUGGGCUCCAGUGGGCCCAGCUCUC .------------((---(((.((((((.(((((..............)))))))))))((((....)))))))))((((((....))))))....... ( -37.24) >DroGri_CAF1 96635 84 - 1 C------------AC---AUUUGGCUACACAUCAUACGCUCUACCCGCUAAUGGCUGCCGCCCAGCUGGGUUACGUGAGCUCCAGUGGGCCCAGCUCGC .------------.(---(((.(((.....................)))))))((((..(((((.(((((...(....).)))))))))).)))).... ( -24.70) >DroYak_CAF1 56832 84 - 1 C------------CC---ACCAGUUGCCCCACCCAUCGCUCUACCCGCUCAUGGCCGCCGCCCAACUGGGCUAUGCCGGCUCCAGCGGACCCAGUUCAC .------------..---...................(((....(((((...((((((.((((....))))...).)))))..)))))....))).... ( -23.30) >DroMoj_CAF1 105164 84 - 1 C------------GC---AUCUGCCGCCCCAUCAUUCGCUCUAUCCGCUAAUGGCCGCCGCCCAGCUGGGCUAUGUGGGCUCCAGUGGGCCCAGUUCGC .------------((---..(((..(((((.(((((.((.......)).)))))((((.((((....))))...))))......).)))).)))...)) ( -26.50) >DroAna_CAF1 68492 87 - 1 C------------CCAUCACCAGCUGCCCCACCCGUCACUCUACCCGCUGAUGGCAGCCGCCCAACUGGGCUACGCCGGCUCCAGUGGACCCAGCUCCC .------------........(((((..(((((((((((.......).)))))).((((((((....))))......))))...))))...)))))... ( -27.00) >consensus C____________CC___ACCAGCCGCCCCAUCAUUCGCUCUACCCGCUAAUGGCCGCCGCCCAACUGGGCUAUGCCGGCUCCAGUGGACCCAGCUCGC ............................................(((((...((((((.((((....))))...)).))))..)))))........... (-19.77 = -19.05 + -0.72)

| Location | 737,926 – 738,019 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.78 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 737926 93 - 22224390 CGGCAGUUCA------CAUCAUCAUCUGGGCCACGUGGGACACUUGGCCACUGGCCACCC---GCUGCCCCAUCAGUUGCCCC---ACCAGUUGCCCCAUCCAUC .(((((....------..........((((.((.(((((.....(((((...))))))))---)))).))))....(((....---..))))))))......... ( -25.80) >DroSec_CAF1 53759 81 - 1 CGGCAGUUCA------CAUCAUCAUUUGGGCCACGUGGGACACUUGGCCACUGGCCACCC---GCUGCC------------CC---ACCAAUUGCCCCAUCCAUC .(((((((..------..........((((.((.(((((.....(((((...))))))))---)))).)------------))---)..)))))))......... ( -26.64) >DroSim_CAF1 23105 81 - 1 CGGCAGUUCA------CAUCAUCAUUUGGGCCACGUGGGACACUUGGCCACUGGCCACCC---GCUGCC------------CC---ACCAAUUGCCCCAUCCAUC .(((((((..------..........((((.((.(((((.....(((((...))))))))---)))).)------------))---)..)))))))......... ( -26.64) >DroEre_CAF1 60282 81 - 1 CGGCAGUUCA------CAUCAUCAUCUGGGCCACGUGGGACACUUGGCCACUGGCCACCC---GCUGCC------------CC---ACCAGUUGCCCCACCCAUC .(((((....------).........((((.((.(((((.....(((((...))))))))---)))).)------------))---).....))))......... ( -25.20) >DroYak_CAF1 56894 81 - 1 CGGCAGCUCA------CACCAUCAUCUGGGCCACGUGGGUCACUUGGCCACUGGCCACCC---GCUGCC------------CC---ACCAGUUGCCCCACCCAUC .(((((((..------..........((((.((.((((((.....((((...))))))))---)))).)------------))---)..)))))))......... ( -29.04) >DroAna_CAF1 68554 93 - 1 GGGCGGUUCGUCGCACCACCACCAGCUGGGGCAUGUGGCAGCCCACCUCAGUGGUCAUCCCGGUCUGUC------------CCAUCACCAGCUGCCCCACCCGUC (((((((.......)))......((((((.(.(((.(((((.(((((.....)))......)).)))))------------.)))).))))))))))........ ( -32.10) >consensus CGGCAGUUCA______CAUCAUCAUCUGGGCCACGUGGGACACUUGGCCACUGGCCACCC___GCUGCC____________CC___ACCAGUUGCCCCACCCAUC .((((((............((.....))(((((.((((.........))))))))).......)))))).................................... (-18.95 = -18.78 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:59 2006