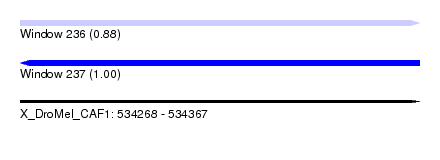
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 534,268 – 534,367 |
| Length | 99 |
| Max. P | 0.998126 |

| Location | 534,268 – 534,367 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 534268 99 + 22224390 ACGAGAUGGUUUUGCAGCACUGCUUCCGACACUUUCAACAUUCUUGCGAAA----------------UACUCUCAAAGG-----UAACGAAAUGUGCCCAGAAAGCGUCGGAACAACCAU .....((((((..(((....)))(((((((.(((((.......(((.((..----------------...)).))).((-----((........))))..))))).))))))).)))))) ( -28.00) >DroSec_CAF1 22310 104 + 1 ACGAGAUGGUUUUGCAGCACUGCUUCCGACACUUUCAACAUUCUUGCGAAA----------------UACUCUCAAAGGUAAGGUAACGAAAUGUGCCCAGAAAGCGUCGGAACAACCAU .....((((((..(((....)))(((((((.(((((...((..(((.((..----------------...)).)))..))..((((........))))..))))).))))))).)))))) ( -29.70) >DroSim_CAF1 22471 104 + 1 ACGAGAUGGUUUUGCAGCACUGCUUCCGACACUUUCAACAUUCUUGCGAAA----------------UACUCUCAAAGGUAAGGUAACGAAAUGUGCCCAGAGAGCGUCGGAACAACCAU .....((((((..(((....)))(((((((.(((((...((..(((.((..----------------...)).)))..))..((((........))))..))))).))))))).)))))) ( -29.80) >DroEre_CAF1 25706 99 + 1 AUGGGAUGGUUUGGCAGCACUGCUUCCGACACUUUCAACAUUCUUGCGAAG----------------UAUUCUCAAAGG-----UAACGAAAUGUGCCCAGAAAGCGUCGGAACAACCAU .....((((((.((((....)))(((((((.(((((.......(((.((..----------------...)).))).((-----((........))))..))))).)))))))))))))) ( -28.00) >DroYak_CAF1 20034 106 + 1 UUGCGAUGGUUU---------GCUUCCGACACUUUCAGCAUUCUUGCGAUGGAAUACCUAGCCGGAACACUCUCAAAGG-----UAACGAAAGGUGCCCAGAAAGCGUCGGAACAACCAU .....((((((.---------(.(((((((.(((((((..((((.((...((....))..)).))))..))......((-----((.(....).))))..))))).)))))))))))))) ( -36.50) >consensus ACGAGAUGGUUUUGCAGCACUGCUUCCGACACUUUCAACAUUCUUGCGAAA________________UACUCUCAAAGG_____UAACGAAAUGUGCCCAGAAAGCGUCGGAACAACCAU .....((((((..(((....)))(((((((.(((((.(((((.(((.........................................)))))))).....))))).))))))).)))))) (-20.51 = -20.84 + 0.33)

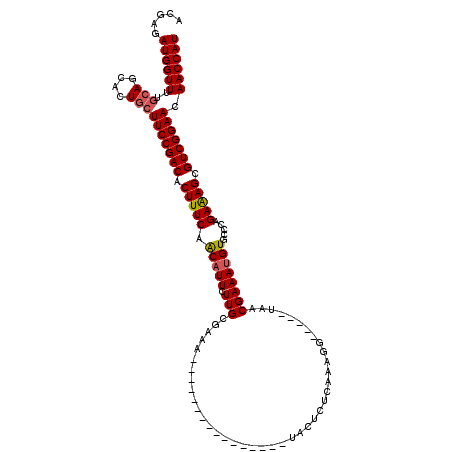
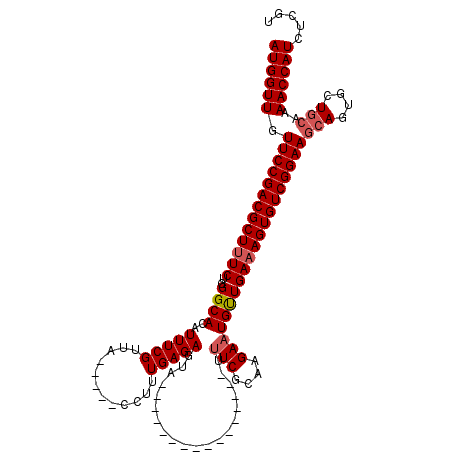
| Location | 534,268 – 534,367 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -30.85 |
| Energy contribution | -31.54 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 534268 99 - 22224390 AUGGUUGUUCCGACGCUUUCUGGGCACAUUUCGUUA-----CCUUUGAGAGUA----------------UUUCGCAAGAAUGUUGAAAGUGUCGGAAGCAGUGCUGCAAAACCAUCUCGU ((((((.(((((((((((((...(((..(((((...-----....)))))...----------------.(((....)))))).)))))))))))))(((....)))..))))))..... ( -34.70) >DroSec_CAF1 22310 104 - 1 AUGGUUGUUCCGACGCUUUCUGGGCACAUUUCGUUACCUUACCUUUGAGAGUA----------------UUUCGCAAGAAUGUUGAAAGUGUCGGAAGCAGUGCUGCAAAACCAUCUCGU ((((((.(((((((((((((...(((........(((((((....)))).)))----------------.(((....)))))).)))))))))))))(((....)))..))))))..... ( -35.50) >DroSim_CAF1 22471 104 - 1 AUGGUUGUUCCGACGCUCUCUGGGCACAUUUCGUUACCUUACCUUUGAGAGUA----------------UUUCGCAAGAAUGUUGAAAGUGUCGGAAGCAGUGCUGCAAAACCAUCUCGU ((((((.((((((((((.((...(((........(((((((....)))).)))----------------.(((....)))))).)).))))))))))(((....)))..))))))..... ( -32.00) >DroEre_CAF1 25706 99 - 1 AUGGUUGUUCCGACGCUUUCUGGGCACAUUUCGUUA-----CCUUUGAGAAUA----------------CUUCGCAAGAAUGUUGAAAGUGUCGGAAGCAGUGCUGCCAAACCAUCCCAU ((((((.(((((((((((((...(((..(((((...-----....)))))...----------------.(((....)))))).)))))))))))))(((....)))..))))))..... ( -33.70) >DroYak_CAF1 20034 106 - 1 AUGGUUGUUCCGACGCUUUCUGGGCACCUUUCGUUA-----CCUUUGAGAGUGUUCCGGCUAGGUAUUCCAUCGCAAGAAUGCUGAAAGUGUCGGAAGC---------AAACCAUCGCAA ((((((.(((((((((((((.((((((.(((((...-----....))))))))))).(((..((....)).((....))..))))))))))))))))..---------.))))))..... ( -41.70) >consensus AUGGUUGUUCCGACGCUUUCUGGGCACAUUUCGUUA_____CCUUUGAGAGUA________________UUUCGCAAGAAUGUUGAAAGUGUCGGAAGCAGUGCUGCAAAACCAUCUCGU ((((((.(((((((((((((..((((..(((((............)))))....................(((....))))))))))))))))))))(((....)))..))))))..... (-30.85 = -31.54 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:13 2006