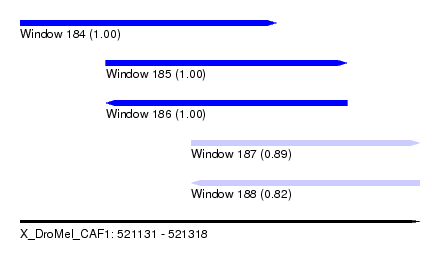
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 521,131 – 521,318 |
| Length | 187 |
| Max. P | 0.999995 |

| Location | 521,131 – 521,251 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -33.72 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 521131 120 + 22224390 UCACCUCGCUAUUUCCCCAGUUUUCUCUUUUUGCUCUGAAGUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUC .......(((((((((((((...............)))..).)))))))))((((((((....((((...(((((..(((((...((....)).)))))..))))))))).)))))))). ( -36.26) >DroSec_CAF1 7341 120 + 1 UCACCUCGCUAUUUCCCCAGUUUUCUCUCUUUGCUCUCAAGUAGAAGUGGCAGUGAAUUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUC .......((((((((...((......))...((((....))))))))))))((((((......((((...(((((..(((((...((....)).)))))..)))))))))...)))))). ( -30.80) >DroSim_CAF1 7734 120 + 1 UCACCUCGCUAUUUCCCCAGUUUUCUCUUUUUGCUCUCAAGUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUC .......(((((((((..((......))....(((....))))))))))))((((((((....((((...(((((..(((((...((....)).)))))..))))))))).)))))))). ( -34.70) >DroEre_CAF1 7286 119 + 1 UCACCUCGCUAU-UCCCCAGUUUUCUCUUUUUGCUCUCAAGUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUC .......(((((-(..(((.((................)).))).))))))((((((((....((((...(((((..(((((...((....)).)))))..))))))))).)))))))). ( -31.49) >DroYak_CAF1 6469 120 + 1 UCACCUCGCCAUUUCCCCAGCUUUCUCUUUUUGCUCUCAAGUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUC .......(((((((((.((((...........))).....).)))))))))((((((((....((((...(((((..(((((...((....)).)))))..))))))))).)))))))). ( -39.00) >consensus UCACCUCGCUAUUUCCCCAGUUUUCUCUUUUUGCUCUCAAGUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUC .......(((((((((.((((...........))).....).)))))))))((((((((....((((...(((((..(((((...((....)).)))))..))))))))).)))))))). (-33.72 = -34.00 + 0.28)

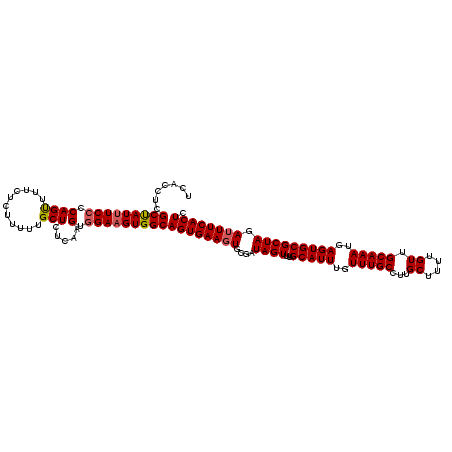
| Location | 521,171 – 521,284 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -34.88 |
| Energy contribution | -35.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.87 |
| SVM RNA-class probability | 0.999995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 521171 113 + 22224390 GUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU------- .(((((((.((((((((((((((.(((((((((((..(((((...((....)).)))))..)))..))))))))....)))))))))).)))).....)))))))........------- ( -36.40) >DroSec_CAF1 7381 113 + 1 GUAGAAGUGGCAGUGAAUUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU------- .........((((((((.(((((.(((((((((((..(((((...((....)).)))))..)))..))))))))....))))).)))).))))(((((........)))))..------- ( -29.80) >DroSim_CAF1 7774 113 + 1 GUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU------- .(((((((.((((((((((((((.(((((((((((..(((((...((....)).)))))..)))..))))))))....)))))))))).)))).....)))))))........------- ( -36.40) >DroEre_CAF1 7325 120 + 1 GUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUCUUUUUGUCCCCU .(((((((.((((((((((((((.(((((((((((..(((((...((....)).)))))..)))..))))))))....)))))))))).)))).....)))))))............... ( -36.40) >DroYak_CAF1 6509 120 + 1 GUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUCUUUUUGUCCCCC .(((((((.((((((((((((((.(((((((((((..(((((...((....)).)))))..)))..))))))))....)))))))))).)))).....)))))))............... ( -36.40) >consensus GUGGAAGUGGCAGUGAAGUGCGAUAGUUUAGCAUUUGUUUGCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU_______ .(((((((.((((((((((((((.(((((((((((..(((((...((....)).)))))..)))..))))))))....)))))))))).)))).....)))))))............... (-34.88 = -35.28 + 0.40)


| Location | 521,171 – 521,284 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 521171 113 - 22224390 -------AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGCAAACAAAUGCUAAACUAUCGCACUUCACUGCCACUUCCAC -------.....((.((((..((((((((...........(((((..........))))).(((((.......))))))))))))).(((.........))).)))).)).......... ( -22.30) >DroSec_CAF1 7381 113 - 1 -------AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGCAAACAAAUGCUAAACUAUCGCAAUUCACUGCCACUUCUAC -------.....((.((((..((((((((...........(((((..........))))).(((((.......))))))))))))).(((.........))).)))).)).......... ( -22.80) >DroSim_CAF1 7774 113 - 1 -------AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGCAAACAAAUGCUAAACUAUCGCACUUCACUGCCACUUCCAC -------.....((.((((..((((((((...........(((((..........))))).(((((.......))))))))))))).(((.........))).)))).)).......... ( -22.30) >DroEre_CAF1 7325 120 - 1 AGGGGACAAAAAGAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGCAAACAAAUGCUAAACUAUCGCACUUCACUGCCACUUCCAC ..((((.....((..((((..((((((((...........(((((..........))))).(((((.......))))))))))))).(((.........))).)))))).....)))).. ( -24.80) >DroYak_CAF1 6509 120 - 1 GGGGGACAAAAAGAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGCAAACAAAUGCUAAACUAUCGCACUUCACUGCCACUUCCAC (((((.........(((....(((((.((((((....((((...(....)...))))...)))))))))))...)))((((......(((.........)))......)))).))))).. ( -27.40) >consensus _______AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGCAAACAAAUGCUAAACUAUCGCACUUCACUGCCACUUCCAC ...............((((..((((((((...........(((((..........))))).(((((.......))))))))))))).(((.........))).))))............. (-21.30 = -21.30 + 0.00)

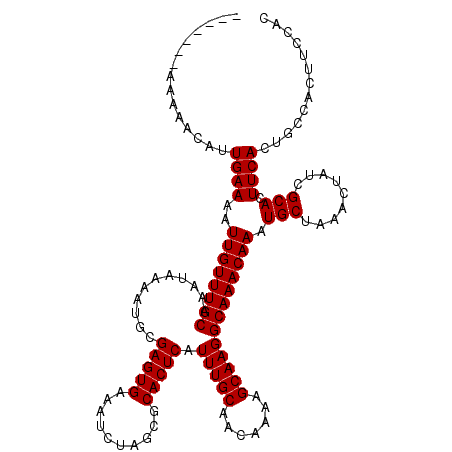
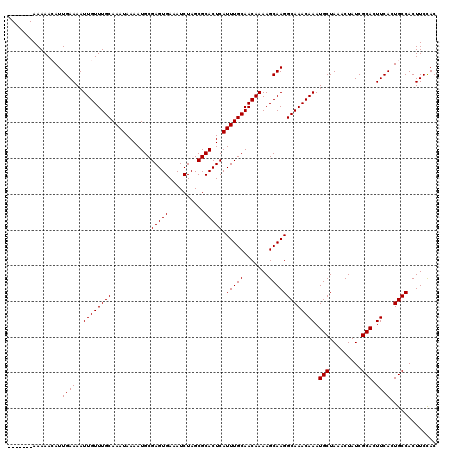
| Location | 521,211 – 521,318 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 521211 107 + 22224390 GCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU-------------CCACUGCGUCCCCCGCUUUUCCAUUUUCCUCUUG ((.(((...((((((((((((((((((..((......)))))).))))))))).)))))....))).))....-------------.....(((.....))).................. ( -19.40) >DroSec_CAF1 7421 107 + 1 GCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU-------------CCACUGCGUCCCCCGCUUUUCCAUUUUCCUCUUG ((.(((...((((((((((((((((((..((......)))))).))))))))).)))))....))).))....-------------.....(((.....))).................. ( -19.40) >DroSim_CAF1 7814 107 + 1 GCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU-------------CCACUGCGUCCCCCGCUUUUCCAUUUUCCUCUUG ((.(((...((((((((((((((((((..((......)))))).))))))))).)))))....))).))....-------------.....(((.....))).................. ( -19.40) >DroEre_CAF1 7365 118 + 1 GCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUCUUUUUGUCCCCUG--CGUCCUCUGCGCUCCCCGCUUUUCCAUUUUCCUCCUG .....((..((((((((((((((((((..((......)))))).))))))))).))))).....................(--(((.....)))).....)).................. ( -21.40) >DroYak_CAF1 6549 120 + 1 GCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUCUUUUUGUCCCCCGCACGUCCCCUGCAUCCCCCGGUUUUCCAUUUUCCUCUUG ((((((...((((((((((((((((((..((......)))))).))))))))).)))))....)))..............(((.......))).......)))................. ( -20.30) >consensus GCCUUGCUUUUGUUGCAAAUGAGUGCGCUAGAUUUCACUCGCAUUUUAUUUGCAAACAAUUUUCAAUGUUUUU_____________CCACUGCGUCCCCCGCUUUUCCAUUUUCCUCUUG ...(((...((((((((((((((((((..((......)))))).))))))))).)))))....))).........................(((.....))).................. (-16.44 = -16.84 + 0.40)

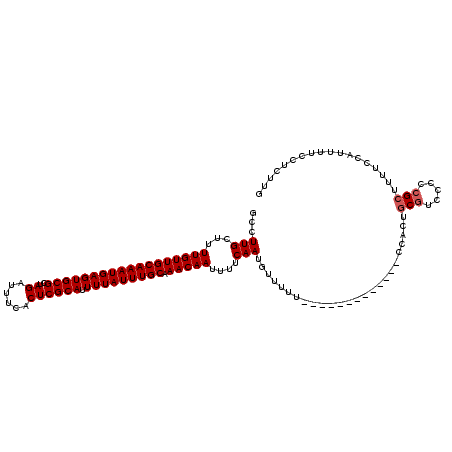
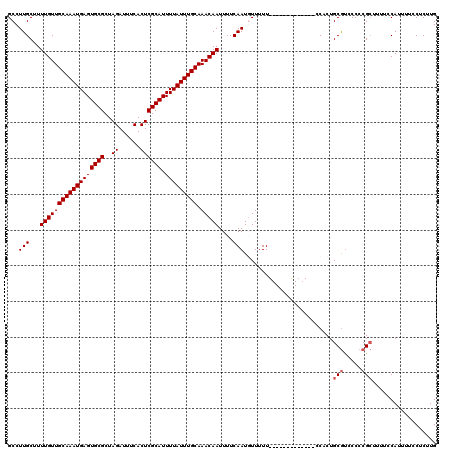
| Location | 521,211 – 521,318 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 521211 107 - 22224390 CAAGAGGAAAAUGGAAAAGCGGGGGACGCAGUGG-------------AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGC ...(((.....(....).(((..(((..(((((.-------------.....)))))....(..((((((.......))))))..)..)))..))).))).(((((.......))))).. ( -22.70) >DroSec_CAF1 7421 107 - 1 CAAGAGGAAAAUGGAAAAGCGGGGGACGCAGUGG-------------AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGC ...(((.....(....).(((..(((..(((((.-------------.....)))))....(..((((((.......))))))..)..)))..))).))).(((((.......))))).. ( -22.70) >DroSim_CAF1 7814 107 - 1 CAAGAGGAAAAUGGAAAAGCGGGGGACGCAGUGG-------------AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGC ...(((.....(....).(((..(((..(((((.-------------.....)))))....(..((((((.......))))))..)..)))..))).))).(((((.......))))).. ( -22.70) >DroEre_CAF1 7365 118 - 1 CAGGAGGAAAAUGGAAAAGCGGGGAGCGCAGAGGACG--CAGGGGACAAAAAGAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGC ..................(((.....))).(((..((--(.(....)....(((((.....(..((((((.......))))))..)))))).)))..))).(((((.......))))).. ( -26.40) >DroYak_CAF1 6549 120 - 1 CAAGAGGAAAAUGGAAAACCGGGGGAUGCAGGGGACGUGCGGGGGACAAAAAGAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGC ...........(((....))).....((((((.((.((((((....)....(((((.....(..((((((.......))))))..))))))..))))))).))))))............. ( -25.90) >consensus CAAGAGGAAAAUGGAAAAGCGGGGGACGCAGUGG_____________AAAAACAUUGAAAAUUGUUUGCAAAUAAAAUGCGAGUGAAAUCUAGCGCACUCAUUUGCAACAAAAGCAAGGC ...(((.....((((...(((.....)))................................(..((((((.......))))))..)..)))).....))).(((((.......))))).. (-18.96 = -19.00 + 0.04)

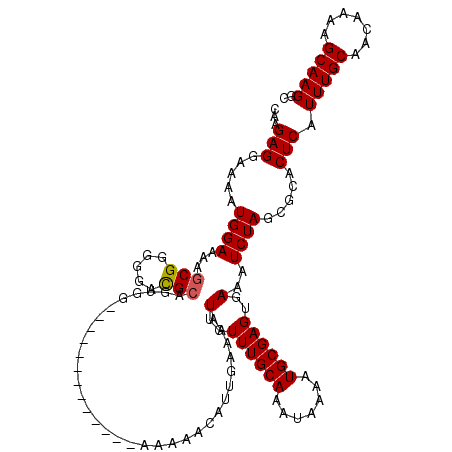
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:43 2006