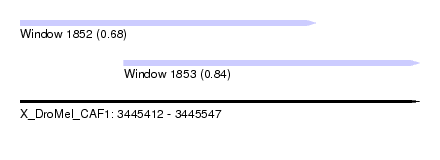
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,445,412 – 3,445,547 |
| Length | 135 |
| Max. P | 0.835464 |

| Location | 3,445,412 – 3,445,512 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -19.40 |
| Energy contribution | -21.04 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3445412 100 + 22224390 AAAGGGUUCCACAACUGAGAUUGCUGGAGAUUGCU-AGGUGUACGGCACCAAAACCAAAUGG--GGGCUUAAAGGGCUUCAAAAUGGG----------GCCUGGAUGUUUUUA ...((.((((((((......))).)))))......-.((((.....))))....))...(((--((((.....(((((((.....)))----------))))....))))))) ( -26.10) >DroSec_CAF1 17671 99 + 1 AAAGGGUUCCAGAACUGAGAUUGCAGGA-----CU-AGGUGUACGGCACC------AAAUGG--GGGCUUAAAGGGCUUCAAAAUGGGGCCCAAAGGGGCCUGGAUUUUGUUU ...(((((((((..(((......)))..-----))-.((((.....))))------......--.........(((((((.....)))))))...)))))))........... ( -32.10) >DroSim_CAF1 17440 99 + 1 AAAGGGUUCCAGAACUGAGAUUGCAGGA-----CU-AGGUGUACGGCACC------AAAUGG--GGGCUUAAAGGGCUUCAAAAUGGGGCCCAAAGGGGCCUGGAUUUUGUUU ...(((((((((..(((......)))..-----))-.((((.....))))------......--.........(((((((.....)))))))...)))))))........... ( -32.10) >DroEre_CAF1 20161 97 + 1 AAAGGGUUCCAGAACCGAGAUUGCAGGA-----CU-AGGUGUACGGCACC------AAAUGGGGGGGCUUAAAAGGCUUGAAAUGAGGGCCCAAAGGGGCCCGGGUUUU---- ...(((((((....((.........)).-----..-.((((.....))))------.......))))))).........(((((..((((((....))))))..)))))---- ( -31.00) >DroYak_CAF1 18938 96 + 1 AAAGGGUUCCCAAACUGAGAUUGCAGAA-----CUGAGGUGCACGGCACC------AAAUGG--AGGCUUAAAGGGCUUUAAAUGGGGGCCCAAAGUUGCCAGGAUUUU---- ...(((((((((..(((......)))..-----....(((((...)))))------.....(--(((((.....))))))...))))))))).................---- ( -31.30) >consensus AAAGGGUUCCAGAACUGAGAUUGCAGGA_____CU_AGGUGUACGGCACC______AAAUGG__GGGCUUAAAGGGCUUCAAAAUGGGGCCCAAAGGGGCCUGGAUUUU_UU_ .......(((...........................(((((...)))))..............((((((...(((((((.....)))))))....)))))))))........ (-19.40 = -21.04 + 1.64)

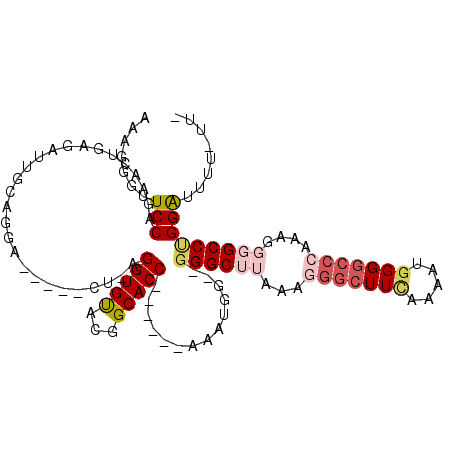
| Location | 3,445,447 – 3,445,547 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -16.79 |
| Energy contribution | -18.19 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3445447 100 + 22224390 AGGUGUACGGCACCAAAACCAAAUGG--GGGCUUAAAGGGCUUCAAAAUGGG----------GCCUGGAUGUUUUUAUUUUUUUUUCGAAACACAGAUUC-----GAGGUUGCUUUG .((((.....))))..((((((((((--((((.....(((((((.....)))----------))))....)))))))))).....(((((.......)))-----))))))...... ( -28.90) >DroSec_CAF1 17701 101 + 1 AGGUGUACGGCACC------AAAUGG--GGGCUUAAAGGGCUUCAAAAUGGGGCCCAAAGGGGCCUGGAUUUUGUUUUUUUUUU---UUAACACAGAUUC-----GAGGUUGCUUUG .((((.....))))------......--.........(((((((.....)))))))((((.(((((.((.(((((.((......---..)).))))).))-----.))))).)))). ( -34.90) >DroSim_CAF1 17470 100 + 1 AGGUGUACGGCACC------AAAUGG--GGGCUUAAAGGGCUUCAAAAUGGGGCCCAAAGGGGCCUGGAUUUUGUUUUUU-UUU---UCAACACAGAUUC-----GAGGUUGCUUUG .((((.....))))------......--.........(((((((.....)))))))((((.(((((.((.(((((.((..-...---..)).))))).))-----.))))).)))). ( -33.90) >DroEre_CAF1 20191 105 + 1 AGGUGUACGGCACC------AAAUGGGGGGGCUUAAAAGGCUUGAAAUGAGGGCCCAAAGGGGCCCGGGUUUU------UUGUUUUCGAAACACAAGCCCACGAGGAGGUUGCUUUG .((((.....))))------....((((.(((((....((((((......((((((....))))))..(((((------........))))).)))))).......))))).)))). ( -37.20) >DroYak_CAF1 18969 95 + 1 AGGUGCACGGCACC------AAAUGG--AGGCUUAAAGGGCUUUAAAUGGGGGCCCAAAGUUGCCAGGAUUUU------U---UUUCGAAACGCAGAUUC-----GAGGUUGCUUUG .(((((...)))))------...(((--..((((...(((((((.....))))))).))))..))).......------.---(((((((.......)))-----))))........ ( -27.60) >consensus AGGUGUACGGCACC______AAAUGG__GGGCUUAAAGGGCUUCAAAAUGGGGCCCAAAGGGGCCUGGAUUUU_UU_UUUUUUUUUCGAAACACAGAUUC_____GAGGUUGCUUUG ........((((((..............((((((...(((((((.....)))))))....))))))((((((......................)))))).......)).))))... (-16.79 = -18.19 + 1.40)

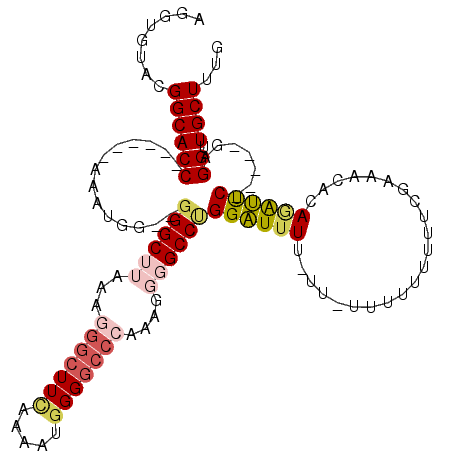
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:37 2006