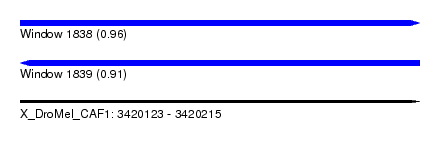
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,420,123 – 3,420,215 |
| Length | 92 |
| Max. P | 0.961013 |

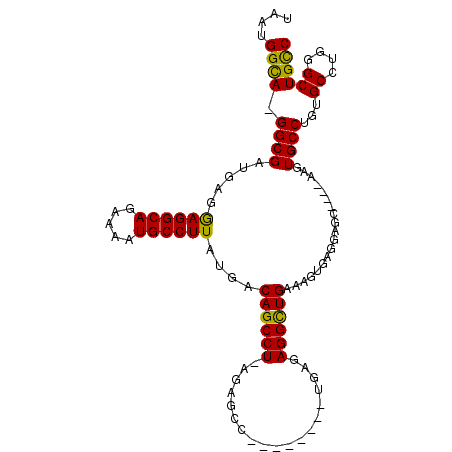
| Location | 3,420,123 – 3,420,215 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -20.77 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
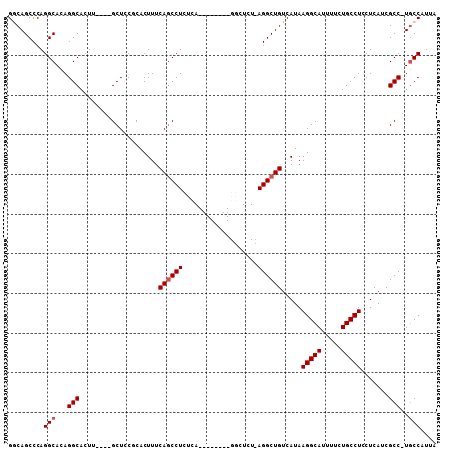
>X_DroMel_CAF1 3420123 92 + 22224390 GACAGCCCAGGCAUAGGCACUUGCUUGCUCCGCACUUUCAGCCUCUC----------------AGGCUGUCAUAAGGCAUUUUCUGCCUUCUCAUCGCCGUGCCAUUA .........(((((.(((...(((.......)))....(((((....----------------.)))))....((((((.....))))))......)))))))).... ( -28.60) >DroSec_CAF1 66120 87 + 1 GGCAGCCCAGGCACAGGCACUU----GCUCCGCACUUUCAGCCUCUC----------------AGGCUGUCAUAAGGCAUUUUCUGCCUCCUCAUCGCC-UGCCAUUG (((.((....))..((((....----............(((((....----------------.))))).....(((((.....))))).......)))-)))).... ( -28.00) >DroSim_CAF1 68194 95 + 1 GGCAGCCCAGGCACAGGCACUU----GCUCCUCACUUUCAGCCUCUCA--------GGCUCUCAGGCUGUCAUAAGGCAUUUUCUGCCUCCUCAUCGCC-UGCCAUUA (((.((....))..((((....----............((((((....--------.......)))))).....(((((.....))))).......)))-)))).... ( -27.50) >DroEre_CAF1 77912 103 + 1 GACAGCCCAGGCACAGGCACUU----GCUCCUCACUUUCAGCCUCUCAUGCUCUCGGGCUCUCAGGCUGUCAUAAGGCAUUUUCUGCCUCCUCAUCGCC-UGCCAUUA ((.((((((((((.((((....----..............))))....))).)).))))).))((((.((.(..(((((.....)))))..).)).)))-)....... ( -29.87) >DroYak_CAF1 68876 95 + 1 GGCAGCCCAGGCACAGGCACUU----GCUUCCCACUUUCAACCUCUCA--------GGCUCUGAGGCUGUCAUAAGGCAUUUUCUGCCUCCUCAUCGCC-UACCAUUA ((((((((((((..((((....----))))............(.....--------))).))).)))))))...(((((.....)))))..........-........ ( -25.70) >consensus GGCAGCCCAGGCACAGGCACUU____GCUCCGCACUUUCAGCCUCUCA________GGCUCU_AGGCUGUCAUAAGGCAUUUUCUGCCUCCUCAUCGCC_UGCCAUUA .........(((...(((....................((((((...................)))))).....(((((.....))))).......)))..))).... (-20.77 = -21.17 + 0.40)

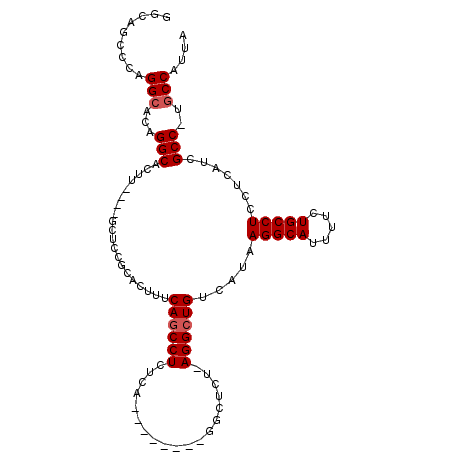
| Location | 3,420,123 – 3,420,215 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -25.97 |
| Energy contribution | -25.25 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3420123 92 - 22224390 UAAUGGCACGGCGAUGAGAAGGCAGAAAAUGCCUUAUGACAGCCU----------------GAGAGGCUGAAAGUGCGGAGCAAGCAAGUGCCUAUGCCUGGGCUGUC ....(((((.((..((..((((((.....))))))....(((((.----------------....)))))......))..))......)))))...((....)).... ( -29.80) >DroSec_CAF1 66120 87 - 1 CAAUGGCA-GGCGAUGAGGAGGCAGAAAAUGCCUUAUGACAGCCU----------------GAGAGGCUGAAAGUGCGGAGC----AAGUGCCUGUGCCUGGGCUGCC ....((((-(.(......((((((.....))))))....(((((.----------------....)))))...(..(((.((----....)))))..)...).))))) ( -35.10) >DroSim_CAF1 68194 95 - 1 UAAUGGCA-GGCGAUGAGGAGGCAGAAAAUGCCUUAUGACAGCCUGAGAGCC--------UGAGAGGCUGAAAGUGAGGAGC----AAGUGCCUGUGCCUGGGCUGCC ....((((-(.(.(.(.(.(((((.......((((((..((((((.......--------....))))))...))))))...----...))))).).).).).))))) ( -34.52) >DroEre_CAF1 77912 103 - 1 UAAUGGCA-GGCGAUGAGGAGGCAGAAAAUGCCUUAUGACAGCCUGAGAGCCCGAGAGCAUGAGAGGCUGAAAGUGAGGAGC----AAGUGCCUGUGCCUGGGCUGUC ......((-(((......((((((.....))))))......)))))..(((((.((.((((...((((.....((.....))----....)))))))))))))))... ( -38.30) >DroYak_CAF1 68876 95 - 1 UAAUGGUA-GGCGAUGAGGAGGCAGAAAAUGCCUUAUGACAGCCUCAGAGCC--------UGAGAGGUUGAAAGUGGGAAGC----AAGUGCCUGUGCCUGGGCUGCC ....((((-(.(.(.(.(.(((((.....((((((((..(((((((......--------...)))))))...)))))..))----)..))))).).).).).))))) ( -29.90) >consensus UAAUGGCA_GGCGAUGAGGAGGCAGAAAAUGCCUUAUGACAGCCU_AGAGCC________UGAGAGGCUGAAAGUGAGGAGC____AAGUGCCUGUGCCUGGGCUGCC ....((((.((((.....((((((.....))))))....((((((...................))))))...................))))...((....)))))) (-25.97 = -25.25 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:24 2006