| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 395,713 – 395,808 |
| Length | 95 |
| Max. P | 0.915358 |

| Location | 395,713 – 395,808 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.82 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -7.88 |
| Energy contribution | -7.85 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 395713 95 - 22224390 UUCUG-GGGAUAUCGCGGCAGAUAAAUUUCAUUAUCUGCAAACGCA-GUCAUAA-AA-UCUGACACCCUAUCCAGUU-GGCAACAAACGGCGGAGGGCUC----------------- (((((-(((.....(((((((((((......))))))))...))).-((((...-..-..)))).)))...((..((-(....)))..))))))).....----------------- ( -29.30) >DroPse_CAF1 8849 89 - 1 AUCUCUGUGAUGUCUGUGCGGAUAAAUUUCAUUAUCUGCAACCGCGACACAUAA-AAAAAAGACAUCAAAUCAAUUUCUGUAACAAAC----------UC----------------- .......((((((((.(((((((((......)))))))))..............-.....))))))))....................----------..----------------- ( -19.05) >DroEre_CAF1 7434 96 - 1 UUCUG-GGGCUGUCGCGGCAGAUAAAUUUCAUUAUCUGCAAACGCA-GUCAUAA-AA-UCUGACAUCGCAUCCAUUCUUGCAACAAACGGCGGAGGGCUC----------------- .....-(((((.((...((((((((......))))))))...(((.-((((...-..-..))))...(((........)))........))))).)))))----------------- ( -29.60) >DroWil_CAF1 13582 114 - 1 CUAUG-AGCUUGUCUCUAUAUAUAAAUUUCAUUAUCUGCAAACGCG-GUCAUAAAAA-UUUGACAUCAAGUCAAUUUUUGUAACAAACGGCCAACUGUUUCGUUCACCCUCCUCUUG ...((-(((..........................((((....)))-)..(((((((-((.(((.....))))))))))))...((((((....)))))).)))))........... ( -18.10) >DroYak_CAF1 2153 96 - 1 UUGUG-GGGCUGUCGCGGCAGAUAAAUUUCAUUAUCUGCAAACGCA-GUCAUAA-AA-UCUGACAUCGCAUCCAUUUUUGCAACAAACGGCGGCGGCCUC----------------- ....(-((((((((((.((((((((......))))))))...((..-((((...-..-..))))...(((........)))......)))))))))))))----------------- ( -37.10) >DroAna_CAF1 11890 90 - 1 AGCUGUGGACUGUCGGCGCGGAUAAAUUUCAUUAUCUGCAAACGCG-CUCAUAA-AA-UCUAACAUCGAAUCAAUUUCUGCAACAGAGGGCAA------------------------ .((((........))))((((((((......))))))))......(-(((....-..-((.......)).......((((...))))))))..------------------------ ( -20.40) >consensus UUCUG_GGGCUGUCGCGGCAGAUAAAUUUCAUUAUCUGCAAACGCA_GUCAUAA_AA_UCUGACAUCGAAUCAAUUUUUGCAACAAACGGCGGAGGGCUC_________________ .................((((((((......)))))))).............................................................................. ( -7.88 = -7.85 + -0.03)

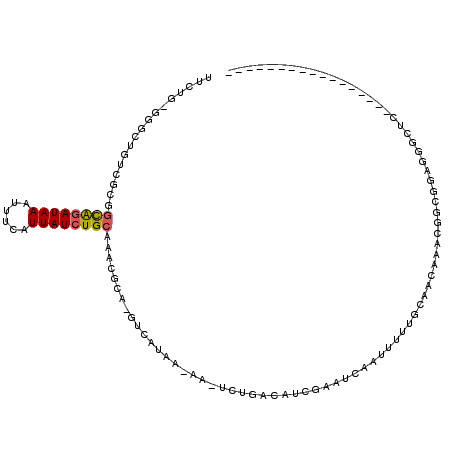
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:19 2006