RNAstrand: reading direction of structured RNAs in multiple sequence alignments
|
RNAstrand predicts the reading direction of a structured RNA in
a multiple sequence alignment by employing a support vector machine (SVM): |
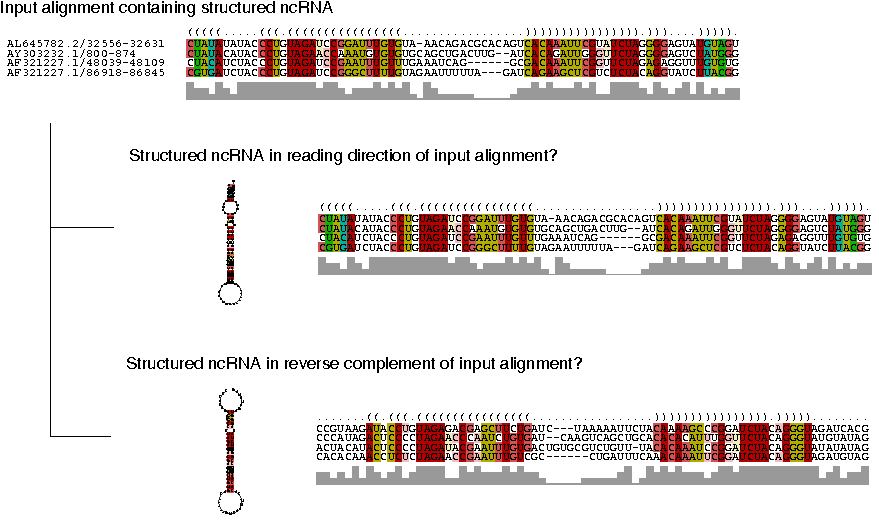
Reading strand information is given in the small asymmetry in the energy rules and in particular by GU pairs
that map to a non-canonical AC pair in the reverse complement of the
alignment. Those asymmetries are utilized by four classification
variables: The energetic differences between strands are captured by
the differences of z-scores and mean folding energies, while the
differences in structure conservation are described by the differences
in structural conservation (SCI) and consensus energies. The output is
a score D which converges to +1 the larger the evidence that RNA is in
reading direction of input alignment and to -1 the larger the evidence
that RNA is in reverse complement of input alignment. Classification
is reported according to a threshold value c:
|
Contact:
Updated:
29 May 2007