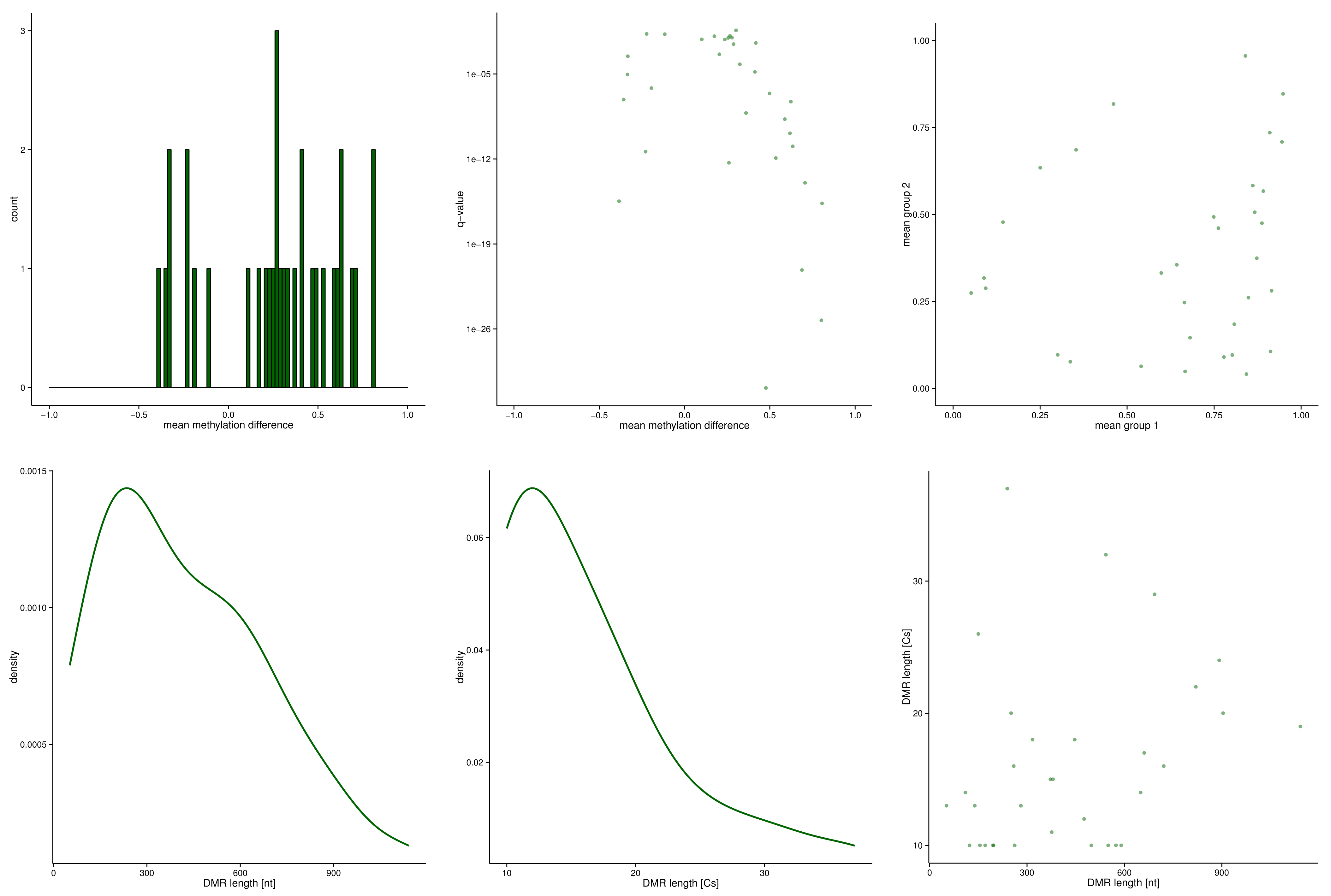
Example: DMRs¶
Example: BAT_DMRcalling¶
Command¶
BAT_DMRcalling -q data/example_metilene_control_case.txt -o DMRs/metilene -a control -b case
Output files¶
DMRs/metilene.out
DMRs/metilene_qval.0.05.out
DMRs/metilene_qval.0.05.bedgraph
DMRs/metilene_qval.0.05.bed
DMRs/metilene_qval.0.05.pdf
View¶
$ head -3 DMRs/metilene.out
chr19 10624503 10624742 3.3e-16 -0.383986 37 4.3e-14 1.5e-19 0.25041 0.63439
chr19 10625238 10625388 6.9e-07 -0.194231 26 4.9e-14 3.1e-10 0.09375 0.28798
chr19 10625393 10625445 9.1e-06 -0.334423 13 4.6e-12 4.1e-09 0.14346 0.47788
$ head -3 DMRs/metilene_qval.0.05.out
chr19 10624503 10624742 3.3e-16 -0.383986 37 0.25041 0.63439
chr19 10625238 10625388 6.9e-07 -0.194231 26 0.09375 0.28798
chr19 10625393 10625445 9.1e-06 -0.334423 13 0.14346 0.47788
$ head -3 DMRs/metilene_qval.0.05.bedgraph
chr19 10624503 10624742 -0.383986
chr19 10625238 10625388 -0.194231
chr19 10625393 10625445 -0.334423
$ head -3 DMRs/metilene_qval.0.05.bed
chr19 10624503 10624742 DMR_1 hypomethylated
chr19 10625238 10625388 DMR_2 hypomethylated
chr19 10625393 10625445 DMR_3 hypomethylated
DMRs/metilene_qval.0.05.pdf
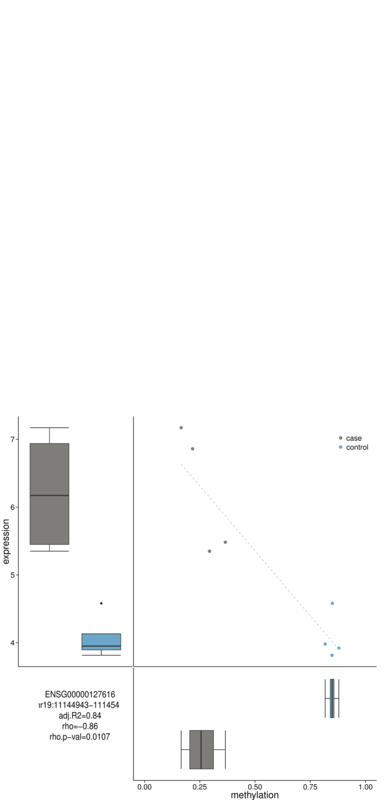
Example: BAT_correlating ¶
Commands¶
Create input for BAT_correlating
bedtools intersect -wa -wb -a DMRs/metilene_qval.0.05.bed -b genomes/hg19/gene.bed | cut -f1-3,9 >DMRs/DMR_gene.txt
ls data/*.bw | grep -v "mean" | grep -v "diff" | sed 's/\t/\n/' >DMRs/methylation_files.list
ls expression/* | sed 's/\t/\n/' >DMRs/expression_files.list
echo -e 'S1\tcontrol\nS2\tcontrol\nS3\tcontrol\nS4\tcontrol\nS5\tcase\nS6\tcase\nS7\tcase\nS8\tcase' >DMRs/sample_to_group.txt
Run BAT_correlating:
BAT_correlating -b DMRs/DMR_gene.txt -e DMRs/expression_files.list -m DMRs/methylation_files.list -g DMRs/sample_to_group.txt -i control,case -o DMRs/correlation
Output files¶
...
correlation.txt
DMRs/correlation_chr19:11144943-11145419_ENSG00000127616.pdf
...
View¶
$ head -3 correlation.txt
plot.pdf ENSG_ID methylation_region adj.R2 rho rho.p-val
DMRs/correlation_chr19:10858170-10858760_ENSG00000079805.pdf ENSG00000079805 chr19:10858170-10858760 0.34 0.83 0.0114
DMRs/correlation_chr19:10928642-10928813_ENSG00000079805.pdf ENSG00000079805 chr19:10928642-10928813 0.69 0.85 0.0075
DMRs/correlation_chr19:10625244-10625340_ENSG00000180739.pdf