| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,452,471 – 10,452,563 |
| Length | 92 |
| Max. P | 0.956078 |

| Location | 10,452,471 – 10,452,563 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 68.29 |
| Shannon entropy | 0.64431 |
| G+C content | 0.48899 |
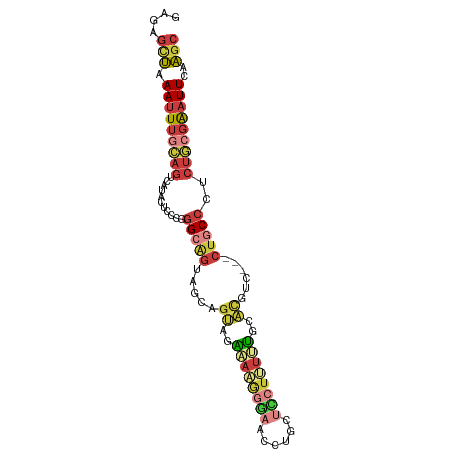
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -12.05 |
| Energy contribution | -13.75 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.956078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
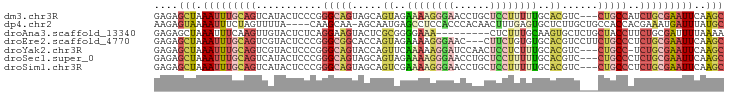
>dm3.chr3R 10452471 92 - 27905053 GAGAGCUAAAUUUGCAGUCAUACUCCCGGGCAGUAGCAGUAGAAAAGGGAACCUGCUCCUUUUUGCACGUC---CUGCCAUCUGCGAAUUCAAGC ....(((.(((((((((...........(((((..((.((..((((((((......))))))))..)))).---)))))..)))))))))..))) ( -26.32, z-score = -0.95, R) >dp4.chr2 19936303 90 + 30794189 AAGAGUAAAAUUUCUAGUUUUA----CAAGCAA-AGCAAUGAGCCUCCACCCACAACUUUGAGUGCUCUUGCUGCCACCACGAAAUGAUUUAUGC ....(((((((.....))))))----)..(((.-.((((.(((((((.............))).))))))))))).................... ( -18.32, z-score = -1.39, R) >droAna3.scaffold_13340 22544655 86 + 23697760 GAGAGCUAAAUUUCAAGUUGUACUCUCAGGAAGUACUCGCGGGGAAA---------CUCUUUGCAAGUGCUCUGCUACCUUCUGCGAUUUUAAAA (((((...(((.....)))...))))).(((.(((((.((((((...---------..)))))).))))))))((........)).......... ( -22.30, z-score = -1.01, R) >droEre2.scaffold_4770 9986021 92 - 17746568 GAGAGCUAAAUUUGCAGUCGUACUCCCGGGCGGCACCAGUAGAAAAGGGAAC---CUUCUGUGUGCACGUCCUUCUGCCCUCUGCGAAUUCAAGC ....(((.(((((((((..(((.....((((((((((((.....((((...)---)))))).)))).)))))...)))...)))))))))..))) ( -29.90, z-score = -1.43, R) >droYak2.chr3R 12850771 91 + 28832112 GAGAGCUAAAUUUGCAGUCGUACUCCCGGGCAGUACCAGUUCAAAAAGGAUCCAACUCCUCUUUGCACGUC---CUGCC-UCUGCGAAUUCAAGC ....(((.(((((((((..........((((((.((..((.((((.((((......)))).)))).)))).---)))))-))))))))))..))) ( -27.20, z-score = -2.73, R) >droSec1.super_0 9535863 92 + 21120651 GAGAGCUAAAUUUGCAGUCAUACUCCCGGGCAGUAGCAGUAGAAAAGGGAACCUGCUCCUUUUUGCACGUC---CUGCCCUCUGCGAAUUCAAGC ....(((.(((((((((..........((((((..((.((..((((((((......))))))))..)))).---)))))).)))))))))..))) ( -28.70, z-score = -1.53, R) >droSim1.chr3R 16506318 92 + 27517382 GAGAGCUAAAUUUGCAGUCAUACUCCCGGGCAGUAGCAGUCGAAAAGGGAACCUGCUCCUUUUUGCACGUC---CUGCCCUCUGCGAAUUCAAGC ....(((.(((((((((..........((((((..((.(((((((((((.......))))))))).)))).---)))))).)))))))))..))) ( -30.10, z-score = -1.97, R) >consensus GAGAGCUAAAUUUGCAGUCAUACUCCCGGGCAGUAGCAGUAGAAAAGGGAACCUGCUCCUUUUUGCACGUC___CUGCCCUCUGCGAAUUCAAGC ....(((.(((((((((...........(((((.....((..((((((((......))))))))..))......)))))..)))))))))..))) (-12.05 = -13.75 + 1.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:26 2011