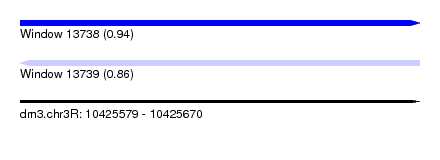
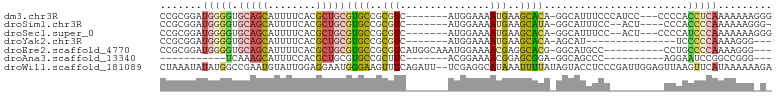
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,425,579 – 10,425,670 |
| Length | 91 |
| Max. P | 0.935519 |

| Location | 10,425,579 – 10,425,670 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 58.97 |
| Shannon entropy | 0.77259 |
| G+C content | 0.55780 |
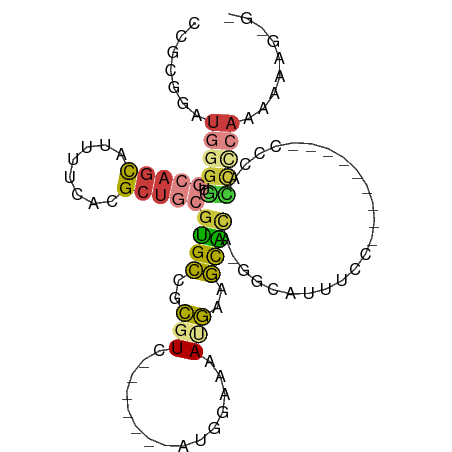
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -9.94 |
| Energy contribution | -12.17 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
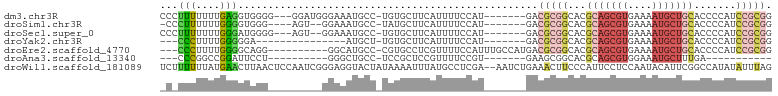
>dm3.chr3R 10425579 91 + 27905053 CCCUUUUUUUGAGGUGGGG---GGAUGGGAAAUGCC-UGUGCUUCAUUUUCCAU-------GACGCGGCACGCAGCGUGAAAAUGCUGCACCCCAUCCGCGG ((((((....)))).))(.---(((((((...((((-.(((..((((.....))-------))))))))).(((((((....)))))))..))))))).).. ( -40.50, z-score = -3.02, R) >droSim1.chr3R 16478479 87 - 27517382 -CCCUUUUUUGGGGUGGG----AGU--GGAAAUGCC-UAUGCUUCAUUUUCCAU-------GACGCGGCACGCAGCGUGAAAAUGCUGCACCCCAUCCGCGG -..........(((((((----.((--.....((((-...((.((((.....))-------)).)))))).(((((((....)))))))))))))))).... ( -35.60, z-score = -2.53, R) >droSec1.super_0 9509048 89 - 21120651 CCCUUUUUUUGGGAUGGGG---AGU--GGAAAUGCC-UGUGCUUCAUUUUCCAU-------GACGCGGCACGCAGCGUGAAAAUGCUGCACCCCAUCCGCGG ...........((((((((---...--.....((((-.(((..((((.....))-------))))))))).(((((((....))))))).)))))))).... ( -39.00, z-score = -3.36, R) >droYak2.chr3R 12823503 76 - 28832112 ---CCCUUUUGGGGGA---------------AUGCU-UGUGCUUCAUUUUCCAU-------GACGCGGCACGCAGCGUGAAAAUGCUGCACCCCAUCCGCGG ---(((...(((((..---------------.....-.(((((((((.....))-------))...)))))(((((((....))))))).)))))...).)) ( -30.50, z-score = -2.47, R) >droEre2.scaffold_4770 9959312 88 + 17746568 ---CCCUUUUGGGGCAGG----------GGCAUGCC-CGUGCCUCGUUUUCCAUUUGCCAUGACGCGGCACGCAGCGUGAAAAUGCUGCACCCCAUCCGCGG ---(((...(((((...(----------((....))-)(((((.((((.............)))).)))))(((((((....))))))).)))))...).)) ( -39.82, z-score = -2.18, R) >droAna3.scaffold_13340 566125 70 + 23697760 ---CCCGGCCGGAUUCCU----------GGGCUGCC-UCCGCUCCGUUUUCCGU-------GAAGCGGCACGCAGCGUGGAAAUGCUUUGA----------- ---.(((.((((....))----------))(((((.-.((((((((.....)).-------).)))))...))))).)))...........----------- ( -23.70, z-score = 0.43, R) >droWil1.scaffold_181089 3282766 100 + 12369635 UCUUUUUUAUGAACUUAACUCCAAUCGGGAGGUACUAUAAAAUUUAUGCCUCGA--AAUCUGAAACUUCCCAUUCCUCCAAUACAUUCGGCCAUAUAUUUAG .......((((..(......((....))((((((..(......)..))))))..--................................)..))))....... ( -10.10, z-score = 0.70, R) >consensus _C_CUUUUUUGGGGUGGG_________GGAAAUGCC_UGUGCUUCAUUUUCCAU_______GACGCGGCACGCAGCGUGAAAAUGCUGCACCCCAUCCGCGG ...(((....)))..................................................(((((...(((((((....))))))).......))))). ( -9.94 = -12.17 + 2.23)
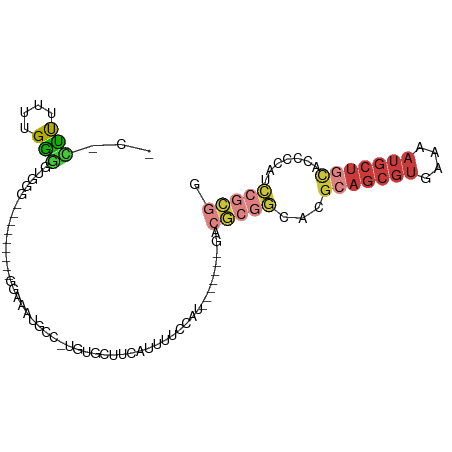
| Location | 10,425,579 – 10,425,670 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 58.97 |
| Shannon entropy | 0.77259 |
| G+C content | 0.55780 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -11.32 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10425579 91 - 27905053 CCGCGGAUGGGGUGCAGCAUUUUCACGCUGCGUGCCGCGUC-------AUGGAAAAUGAAGCACA-GGCAUUUCCCAUCC---CCCCACCUCAAAAAAAGGG ....((((((((.(((((........)))))(((((((.((-------((.....)))).))...-))))).))))))))---.(((............))) ( -34.70, z-score = -2.19, R) >droSim1.chr3R 16478479 87 + 27517382 CCGCGGAUGGGGUGCAGCAUUUUCACGCUGCGUGCCGCGUC-------AUGGAAAAUGAAGCAUA-GGCAUUUCC--ACU----CCCACCCCAAAAAAGGG- ((.....(((((((((((........)))))(((((((.((-------((.....)))).))...-)))))....--...----...)))))).....)).- ( -31.80, z-score = -1.68, R) >droSec1.super_0 9509048 89 + 21120651 CCGCGGAUGGGGUGCAGCAUUUUCACGCUGCGUGCCGCGUC-------AUGGAAAAUGAAGCACA-GGCAUUUCC--ACU---CCCCAUCCCAAAAAAAGGG ..(.((((((((.(((((........)))))(((((((.((-------((.....)))).))...-)))))....--...---))))))))).......... ( -35.60, z-score = -2.78, R) >droYak2.chr3R 12823503 76 + 28832112 CCGCGGAUGGGGUGCAGCAUUUUCACGCUGCGUGCCGCGUC-------AUGGAAAAUGAAGCACA-AGCAU---------------UCCCCCAAAAGGG--- ((.....(((((.(((((........)))))((((....((-------((.....)))).)))).-.....---------------..)))))...)).--- ( -26.80, z-score = -0.93, R) >droEre2.scaffold_4770 9959312 88 - 17746568 CCGCGGAUGGGGUGCAGCAUUUUCACGCUGCGUGCCGCGUCAUGGCAAAUGGAAAACGAGGCACG-GGCAUGCC----------CCUGCCCCAAAAGGG--- ((.....(((((((((((........)))))(((((.((((((.....)))....))).)))))(-((....))----------)..))))))...)).--- ( -37.30, z-score = -0.79, R) >droAna3.scaffold_13340 566125 70 - 23697760 -----------UCAAAGCAUUUCCACGCUGCGUGCCGCUUC-------ACGGAAAACGGAGCGGA-GGCAGCCC----------AGGAAUCCGGCCGGG--- -----------.....((..((((..(((((.(.(((((((-------.........))))))))-.)))))..----------.))))....))....--- ( -26.50, z-score = -0.88, R) >droWil1.scaffold_181089 3282766 100 - 12369635 CUAAAUAUAUGGCCGAAUGUAUUGGAGGAAUGGGAAGUUUCAGAUU--UCGAGGCAUAAAUUUUAUAGUACCUCCCGAUUGGAGUUAAGUUCAUAAAAAAGA .......((((..(..((((.((((((...((((....))))..))--)))).))))..............((((.....))))....)..))))....... ( -15.70, z-score = 0.93, R) >consensus CCGCGGAUGGGGUGCAGCAUUUUCACGCUGCGUGCCGCGUC_______AUGGAAAAUGAAGCACA_GGCAUUUCC_________CCCACCCCAAAAAAG_G_ .......(((((.(((((........)))))((((..(((...............)))..))))........................)))))......... (-11.32 = -11.82 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:25 2011