
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,416,565 – 10,416,667 |
| Length | 102 |
| Max. P | 0.872902 |

| Location | 10,416,565 – 10,416,667 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.43 |
| Shannon entropy | 0.62501 |
| G+C content | 0.44230 |
| Mean single sequence MFE | -17.38 |
| Consensus MFE | -10.10 |
| Energy contribution | -9.77 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
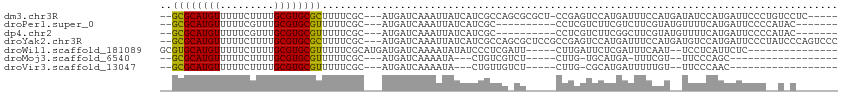
>dm3.chr3R 10416565 102 - 27905053 --GCGCAUGUUUUUCUUUUGCGUGCGCUUUUCGC---AUGAUCAAAUUAUCAUCGCCAGCGCGCU-CCGAGUCCAUGAUUUCCAUGAUAUCCAUGAUUCCCUGUCCUC----- --..(((...........)))(((((((...((.---(((((......)))))))..))))))).-..(((..((.((....((((.....))))..))..))..)))----- ( -23.50, z-score = -1.34, R) >droPer1.super_0 5606529 91 + 11822988 --GCGCAUGUUUUUCGUUUGCGUGCGUUUUUCGC---AUGAUCAAAUUAUCAUCGC----------CCUCGUCUUCGUCUUCGUAUGUUUUCAUGAUUCCCCAUAC------- --((((((((.........)))))))).....((---(((((......))))).))----------................(((((..............)))))------- ( -14.94, z-score = -1.04, R) >dp4.chr2 14224735 91 - 30794189 --GCGCAUGUUUUUCGUUUGCGUGCGUUUUUCGC---AUGAUCAAAUUAUCAUCGC----------CCUCGUCUUCGGCUUCGUAUGUUUUCAUGAUUCCCCAUAC------- --((((((((.........)))))))).....((---(((((......))))).))----------..........((..((((........))))..))......------- ( -17.30, z-score = -0.98, R) >droYak2.chr3R 12813802 108 + 28832112 --GCGCAUGUUUUUCUUUUGCGUGCGCUUUUCGC---AUGAUCAAAUUAUCAUCGCCAGCGCUCCGCCGAGUCCAUGAUUUCCAUGAUGUCCAUGAUUCCCUAUCCCAGUCCC --((((((((.........)))))))).....((---(((((......))))).))((..((((....))))...)).....((((.....)))).................. ( -22.10, z-score = -1.04, R) >droWil1.scaffold_181089 3273502 91 - 12369635 GCGUGCAUGUUUUUCUUUUGCGUGCGUUUUUCGCAUGAUGAUCAAAAUAUAUCCCUCGAUU-----CUUGAUUCUCGAUUUCAAU--UCCUCAUUCUC--------------- ((((((((((.........))))))).....))).(((.((((((.....(((....))).-----.)))))).)))........--...........--------------- ( -14.50, z-score = -0.57, R) >droMoj3.scaffold_6540 3451153 78 + 34148556 --GCGCAUGUUUUUCUUUUGCGUGCGUUUUUCGC---AUGAUCAAAAUA---CUGUCGUCU-----CUUG-UGCAUGA-UUUCGU--UUCCCAGC------------------ --(((((((......(((((((((((.....)))---)))..)))))..---....))...-----..))-)))....-......--........------------------ ( -13.40, z-score = 0.21, R) >droVir3.scaffold_13047 11064564 79 - 19223366 --GCGCAUGUUUUUCUUUUGCGUGCGUUUUUCGC---AUGAUCAAAAUA---CUGUUGUCU-----CUUG-CGCAUGAUUUUUGU--UUCCCAAC------------------ --(((((........(((((((((((.....)))---)))..))))).(---(....))..-----..))-)))...........--........------------------ ( -15.90, z-score = -1.07, R) >consensus __GCGCAUGUUUUUCUUUUGCGUGCGUUUUUCGC___AUGAUCAAAUUAUCAUCGCCG_CU_____CCUGGUCCAUGAUUUCCAU__UUUCCAUGAUUCCC__U_C_______ ..((((((((.........))))))))...................................................................................... (-10.10 = -9.77 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:23 2011