| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,385,760 – 10,385,863 |
| Length | 103 |
| Max. P | 0.987752 |

| Location | 10,385,760 – 10,385,863 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.97 |
| Shannon entropy | 0.70615 |
| G+C content | 0.43156 |
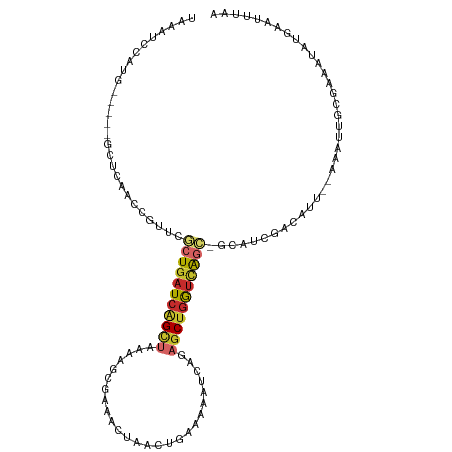
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -9.71 |
| Energy contribution | -10.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
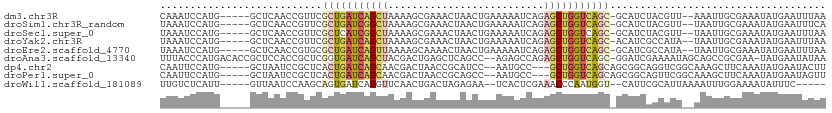
>dm3.chr3R 10385760 103 + 27905053 CAAAUCCAUG-----GCUCAACCGUUCGCUGAUCAGCUAAAAGCGAAACUAACUGAAAAAUCAGAGCUGGUCAGC-GCAUCUACGUU--AAAUUGCGAAAUAUGAAUUUAA .((((.((((-----..((........(((((((((((...((.....))..((((....)))))))))))))))-(((........--....)))))..)))).)))).. ( -28.40, z-score = -3.29, R) >droSim1.chr3R_random 570266 103 + 1307089 UAAAUCCAUG-----GCUCAACCGUUCGCUGAUCGGCUAAAAGCGAAACUAACUGAAAAAUCAGAGCUGGUCAGC-GCAUCUACGUU--UAAUUGCGAAAUAUGAAUUUCA .((((.((((-----..((........(((((((((((...((.....))..((((....)))))))))))))))-(((........--....)))))..)))).)))).. ( -27.00, z-score = -2.34, R) >droSec1.super_0 9469497 103 - 21120651 UAAAUCCAUG-----GCUCAACCGUUCGCUCAUCGGCUAAAAGCGAAACUAACUGAAAAAUCAGAGCUGGUCAGC-GCAUCUACGUU--UAAUUGCGAAAUAUGAAUUUAA (((((.((((-----..((........(((.(((((((...((.....))..((((....))))))))))).)))-(((........--....)))))..)))).))))). ( -22.60, z-score = -1.24, R) >droYak2.chr3R 12781647 103 - 28832112 UAAAUCCAUG-----GCUCAACCGUUCGCUGAUCAGCUAAAAGCGAAACUAACUGAAAAAUCAGAGCUGGUCAGC-ACAUCGCCAUA--UAAUUGCGAAAUAUGAAUUUAA (((((.((((-----............(((((((((((...((.....))..((((....)))))))))))))))-...((((....--.....))))..)))).))))). ( -28.40, z-score = -3.70, R) >droEre2.scaffold_4770 9918393 103 + 17746568 UAAAUCCAUG-----GCUCAACCGUGCGCUGAUCAGUUAAAAGCAAAACUAACUGAAAAAUCAGAGCUGGUCAGC-GCAUCGCCAUA--UAAUUGCGAAAUAUGAAUUUAA (((((.((((-----..((....(((((((((((((((...((.....))..((((....)))))))))))))))-)))).((....--.....))))..)))).))))). ( -32.20, z-score = -4.56, R) >droAna3.scaffold_13340 534120 107 + 23697760 UUUACCCAUGACACCGCUCCACCGCUCGGUGAUCAGCUACGACUGAGCUCAGCC--AGAGCCAGAGCUGGUCAGC-GGAUCGAAAAUAGCAGCCGCGAA-UAUGAAUAUAA ......((((...(((((..(((....)))((((((((....(((.((((....--.))))))))))))))))))-))..........((....))...-))))....... ( -33.40, z-score = -2.12, R) >dp4.chr2 14185948 101 + 30794189 CAAUUCCAUG-----GCUAAUCCGCUCACUGAUCAGCAACGACUAACCGCAUCC--AAUGCC---GCUGGUCAGCAGCGGCAGGUCGGCAAAGCUUCAAAUAUGAAUACUU ..((((...(-----(((...(((((..(((((((((...(......)((((..--.)))).---))))))))).)))))..))))(((...)))........)))).... ( -27.90, z-score = -1.34, R) >droPer1.super_0 5563789 101 - 11822988 CAAUUCCAUG-----GCUAAUCCGCUCACUGAUCAGCAACGACUAACCGCAGCC--AAUGCC---GCUGGUCAGCAGCGGCAGUUCGGCAAAGCUUCAAAUAUGAAUAGUU ..........-----(((.(((........))).)))...(((((...((.(((--..((((---((((.....))))))))....)))...))((((....))))))))) ( -28.10, z-score = -0.97, R) >droWil1.scaffold_181089 3238356 97 + 12369635 UUGUCUCAUU-----GUUAAUCCAAGCAGUGAUCAGGUUCAACUGACUAGAGAA--UCACUCGAAACCCAAUGGU--CAUUCGCAUUAAAAUUUGGAAAAUAUUUC----- .....(((((-----(((......))))))))((((......))))(((((...--.....(((((((....)))--..))))........)))))..........----- ( -17.49, z-score = -0.42, R) >consensus UAAAUCCAUG_____GCUCAACCGUUCGCUGAUCAGCUAAAAGCGAAACUAACUGAAAAAUCAGAGCUGGUCAGC_GCAUCGACAUU__AAAUUGCGAAAUAUGAAUUUAA ...........................(((((((((((..........................))))))))))).................................... ( -9.71 = -10.27 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:20 2011