| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,346,110 – 10,346,204 |
| Length | 94 |
| Max. P | 0.810956 |

| Location | 10,346,110 – 10,346,204 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.94 |
| Shannon entropy | 0.59320 |
| G+C content | 0.46655 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -8.53 |
| Energy contribution | -8.97 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10346110 94 + 27905053 -AUUUUACCACCGAACUGUCAGAAUUUCACGCGAUGCCUGACACUUGAAUAUUACUG--AAUUCCAGC-------CACGUGGAGCCACGCCC-----ACAAUUAUGUCG- -..........(((..((((((.(((......)))..))))))((.((((.......--.)))).)).-------...((((.((...))))-----)).......)))- ( -15.30, z-score = -0.49, R) >droEre2.scaffold_4770 9877942 91 + 17746568 -UUUUCACCACCGAGCUGUCAGAAUUUCACGCGAUGCCUGACACUUGGUUAUUGUUG--AAUUCCAGC-------CACGUGGAG--ACGCC------ACACUUAUACCC- -(((((((.((((((.((((((.(((......)))..))))))))))))....((((--.....))))-------...))))))--)....------............- ( -22.60, z-score = -2.18, R) >droYak2.chr3R 12741116 93 - 28832112 -AUUUUACCACCGAGCUGUCAGAAUUUCACGCGAUGCCUGACACUUGGUUAUUGCUG--AAUUCCAGC-------CACGUGGAG--ACGCCC-----ACGAUUAUUUCCG -........((((((.((((((.(((......)))..))))))))))))....((((--.....))))-------..(((((..--....))-----))).......... ( -27.20, z-score = -3.59, R) >droSec1.super_0 9424694 92 - 21120651 -AUUUUACCACCGAGCUGUCAGAAUUUCACGCGAUGCCUGACACUUGAAUAUUGCUG--AAUUCCAGC-------CAUGUGGAU--ACGCCC-----ACAAUUAUGUCC- -..........((((.((((((.(((......)))..))))))))))......((((--.....))))-------..(((((..--....))-----))).........- ( -21.90, z-score = -2.32, R) >droSim1.chr3R 16406467 92 - 27517382 -AUUUUACCACCGAGCUGUCAGAAUUUCACGCGAUGCCUGACACUUGAAUAUUGCUG--AAUUCCAGC-------CACGUGGAU--ACGCCC-----ACAAUUAUGUCC- -......((((((((.((((((.(((......)))..))))))))))......((((--.....))))-------...))))..--......-----............- ( -21.20, z-score = -2.22, R) >dp4.chr2 14146242 107 + 30794189 UCUGUUCCGGAGGAGCUGUCACAAUUUCACGUAAUGCCUGACACUUGUUCAUUGCGA--AACAACAGCACCACCCCACCCCACCCCACACCCCACUGAGAGUCGUGUCC- ........((.((.(((((.....((((..((((((...(((....)))))))))))--))..))))).)).))............((((..(.......)..))))..- ( -18.30, z-score = 0.96, R) >droWil1.scaffold_181089 12112125 96 - 12369635 ---GUUUCAAUUUAAAUGUUUAUGCUUAAUAUUAUGAGAAAUGCAUACUUAUUAUGGCCACUCGUGGC-------CACGUGCCGAAAUUAGCA---UAGAAAUGUAUUC- ---(((((......((((..(((((.................)))))..)))).((((((....))))-------)).((((........)))---).)))))......- ( -20.83, z-score = -1.46, R) >consensus _AUUUUACCACCGAGCUGUCAGAAUUUCACGCGAUGCCUGACACUUGAUUAUUGCUG__AAUUCCAGC_______CACGUGGAG__ACGCCC_____ACAAUUAUGUCC_ .......((((((((.((((((....((....))...))))))))))......((((.......))))..........))))............................ ( -8.53 = -8.97 + 0.44)

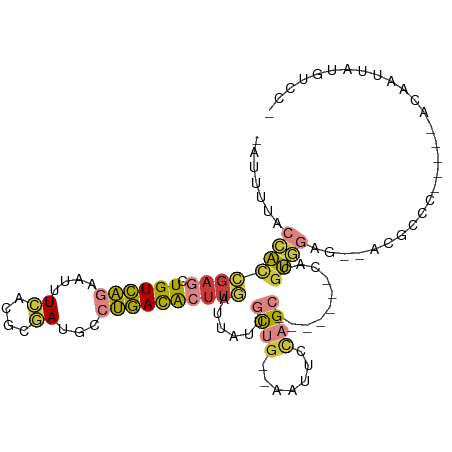

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:18 2011