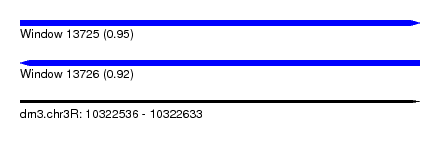
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,322,536 – 10,322,633 |
| Length | 97 |
| Max. P | 0.949258 |

| Location | 10,322,536 – 10,322,633 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Shannon entropy | 0.45551 |
| G+C content | 0.35402 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.04 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10322536 97 + 27905053 -----------------UGCCGUAUGCUGUUUUAUUUUUACCUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGCCUGACUCACGACUU-- -----------------....((..((.((((((((((..((....((((((((((((.....)))).)))))))).....))..)))))))---)..)).))...)).........-- ( -19.30, z-score = -1.20, R) >droSim1.chr3R 16382842 92 - 27517382 ----------------------UGCCGUGUUUUAUUUUUACCUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGCCUGACUCACGACUU-- ----------------------...((((..(((............((((((((((((.....)))).))))))))....((((((......---.))))))..)))..))))....-- ( -20.70, z-score = -2.15, R) >droSec1.super_0 9401280 97 - 21120651 -----------------UGCCGUAUGCUGUUUUAUUUUUACCUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGCCUGACUCACGACUU-- -----------------....((..((.((((((((((..((....((((((((((((.....)))).)))))))).....))..)))))))---)..)).))...)).........-- ( -19.30, z-score = -1.20, R) >droYak2.chr3R 12716670 97 - 28832112 -----------------GGCCGUAUGCUGUUUUAUUUUUACCUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGCCUGACUCACGACUU-- -----------------(((.(((......((((((((..((....((((((((((((.....)))).)))))))).....))..)))))))---).))).))).............-- ( -22.10, z-score = -1.87, R) >droEre2.scaffold_4770 9854238 97 + 17746568 -----------------UGCCGUAUGCUGUUUUAUUUUUACCUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGCCUGACUCACGACUU-- -----------------....((..((.((((((((((..((....((((((((((((.....)))).)))))))).....))..)))))))---)..)).))...)).........-- ( -19.30, z-score = -1.20, R) >droAna3.scaffold_13340 486644 91 + 23697760 ----------------------CGCCGUUUUGUGUCUCU-CCCACAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGCCUGACUCACGACCG-- ----------------------....(((.((.(((...-......((((((((((((.....)))).))))))))....((((((......---.))))))...))).)).)))..-- ( -22.00, z-score = -1.80, R) >dp4.chr2 16382018 114 + 30794189 UGCUGUUUUUUAUACACUUUUGUGGCGUUUUUUUCUGUAUUUUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGACUGACUCACGACUU-- ...................((((((.(((.................((((((((((((.....)))).))))))))...(((((((......---.)))))))..)))))))))...-- ( -23.30, z-score = -1.48, R) >droPer1.super_0 7806849 113 - 11822988 UGCUGUUUUUUAUACACUUUUAUGGC-UCUUUUUCUGUAUUUUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGACUGACUCACGACUU-- ((((((.......)))......((((-..((((..(((((((....)))))))...))))..))))....)))......(((((((......---.)))))))..............-- ( -19.70, z-score = -1.06, R) >droWil1.scaffold_181089 2405908 92 - 12369635 ----------------------CAUUUGUUUAUUUCGUUUUUUUAGAAUUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA---AUUGCCGACUGACUCACGACUU-- ----------------------............((((....((((...(((((((((.....)))).)))))......(((((((......---.)))))))))))...))))...-- ( -18.60, z-score = -1.63, R) >droGri2.scaffold_14830 5380685 91 + 6267026 ----------------------------UGUUUUUGUUUUUUUUUAGAAUGCAAUUGGAGCUGUCAAUUUGCAUUUAAUUCGGUAAAAAUAAUAAAUUGUCGACUGACUCACGACUCAC ----------------------------.....((((((((.....((((((((((((.....)))).))))))))........))))))))......((((.........)))).... ( -13.42, z-score = 0.30, R) >droMoj3.scaffold_6540 16110229 91 - 34148556 -----------------------UGUAUUUCUUUUCUUUUUUUUUAGAGUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCAAAAAUAA---AUUGCCGAUUGACUCACGACUU-- -----------------------.......................((((((((((((.....)))).)))).......(((((((......---.)))))))...)))).......-- ( -18.40, z-score = -1.83, R) >droVir3.scaffold_13047 4628827 75 - 19223366 ---------------------------------------UUUUUUAGAAUGAAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCAAAAAUAA---AUUGCCGACUGACUCACGACUU-- ---------------------------------------.......((((((((((((.....))))))).)))))...(((((((......---.)))))))..............-- ( -15.70, z-score = -2.65, R) >consensus ______________________UAUCCUGUUUUAUUUUUUCCUUUAGAAUGCAAUUGGAACUGUCAAUUUGCAUUUAAUUCGGCGAAAAUAA___AUUGCCGACUGACUCACGACUU__ ..............................................((((((((((((.....)))).))))))))....((((((..........))))))................. (-14.36 = -14.04 + -0.32)

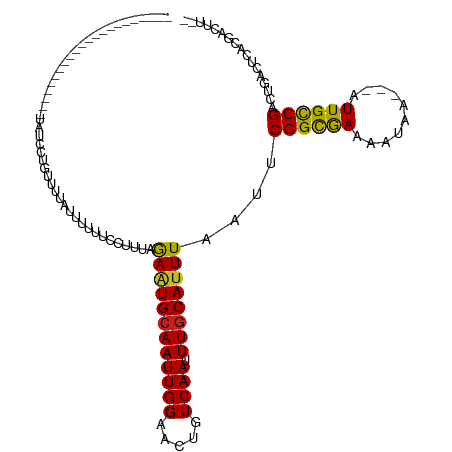
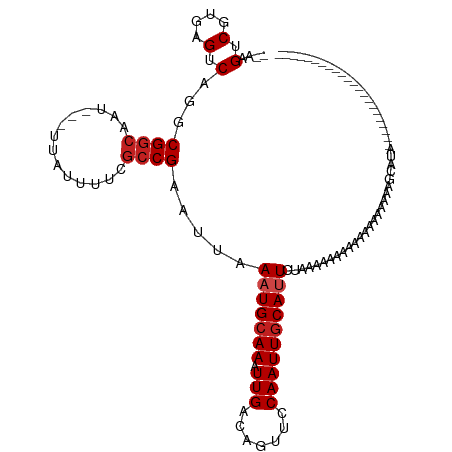
| Location | 10,322,536 – 10,322,633 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Shannon entropy | 0.45551 |
| G+C content | 0.35402 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -11.63 |
| Energy contribution | -12.05 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10322536 97 - 27905053 --AAGUCGUGAGUCAGGCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAGGUAAAAAUAAAACAGCAUACGGCA----------------- --..((((((.(((....)))..(---(((((((..((......(((((((.(((.......)))))))))).....))..))))))))......)))))).----------------- ( -25.30, z-score = -2.93, R) >droSim1.chr3R 16382842 92 + 27517382 --AAGUCGUGAGUCAGGCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAGGUAAAAAUAAAACACGGCA---------------------- --..((((((.(((....)))..(---(((((((..((......(((((((.(((.......)))))))))).....))..)))))))).)))))).---------------------- ( -27.40, z-score = -4.12, R) >droSec1.super_0 9401280 97 + 21120651 --AAGUCGUGAGUCAGGCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAGGUAAAAAUAAAACAGCAUACGGCA----------------- --..((((((.(((....)))..(---(((((((..((......(((((((.(((.......)))))))))).....))..))))))))......)))))).----------------- ( -25.30, z-score = -2.93, R) >droYak2.chr3R 12716670 97 + 28832112 --AAGUCGUGAGUCAGGCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAGGUAAAAAUAAAACAGCAUACGGCC----------------- --..((((((.(((....)))..(---(((((((..((......(((((((.(((.......)))))))))).....))..))))))))......)))))).----------------- ( -24.40, z-score = -2.59, R) >droEre2.scaffold_4770 9854238 97 - 17746568 --AAGUCGUGAGUCAGGCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAGGUAAAAAUAAAACAGCAUACGGCA----------------- --..((((((.(((....)))..(---(((((((..((......(((((((.(((.......)))))))))).....))..))))))))......)))))).----------------- ( -25.30, z-score = -2.93, R) >droAna3.scaffold_13340 486644 91 - 23697760 --CGGUCGUGAGUCAGGCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUGUGGG-AGAGACACAAAACGGCG---------------------- --..(((((.......)))))...---.......(((((.....(((((((.(((.......)))))))))).((((..-.....))))...)))))---------------------- ( -23.20, z-score = -0.67, R) >dp4.chr2 16382018 114 - 30794189 --AAGUCGUGAGUCAGUCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAAAAUACAGAAAAAAACGCCACAAAAGUGUAUAAAAAACAGCA --..((.(((......(((((...---........)))))....(((((((.(((.......))))))))))....................))).))..................... ( -17.10, z-score = -0.33, R) >droPer1.super_0 7806849 113 + 11822988 --AAGUCGUGAGUCAGUCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAAAAUACAGAAAAAGA-GCCAUAAAAGUGUAUAAAAAACAGCA --.....(((..((..(((((...---........)))))....(((((((.(((.......))))))))))..................))-..)))..................... ( -15.80, z-score = 0.06, R) >droWil1.scaffold_181089 2405908 92 + 12369635 --AAGUCGUGAGUCAGUCGGCAAU---UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAAUUCUAAAAAAACGAAAUAAACAAAUG---------------------- --...((((.......(((((...---........)))))......(((((.(((.......))))))))...........))))............---------------------- ( -15.30, z-score = -1.00, R) >droGri2.scaffold_14830 5380685 91 - 6267026 GUGAGUCGUGAGUCAGUCGACAAUUUAUUAUUUUUACCGAAUUAAAUGCAAAUUGACAGCUCCAAUUGCAUUCUAAAAAAAAACAAAAACA---------------------------- ....((((.........)))).......................(((((((.(((.......))))))))))...................---------------------------- ( -10.30, z-score = 0.50, R) >droMoj3.scaffold_6540 16110229 91 + 34148556 --AAGUCGUGAGUCAAUCGGCAAU---UUAUUUUUGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCACUCUAAAAAAAAAGAAAAGAAAUACA----------------------- --.....(((..((..(((((((.---......)))))))......(((((.(((.......))))))))..................))..))).----------------------- ( -18.40, z-score = -2.32, R) >droVir3.scaffold_13047 4628827 75 + 19223366 --AAGUCGUGAGUCAGUCGGCAAU---UUAUUUUUGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUUCAUUCUAAAAAA--------------------------------------- --..(.(....).)..(((((((.---......)))))))....((((.((((((.......))))))))))........--------------------------------------- ( -16.50, z-score = -2.83, R) >consensus __AAGUCGUGAGUCAGGCGGCAAU___UUAUUUUCGCCGAAUUAAAUGCAAAUUGACAGUUCCAAUUGCAUUCUAAAAAAAAAAAAAAAAAAGCAUA______________________ ....(.(....).)...((((..............)))).....(((((((.(((.......))))))))))............................................... (-11.63 = -12.05 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:13 2011