| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,316,091 – 10,316,199 |
| Length | 108 |
| Max. P | 0.998422 |

| Location | 10,316,091 – 10,316,199 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.62 |
| Shannon entropy | 0.39403 |
| G+C content | 0.41740 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
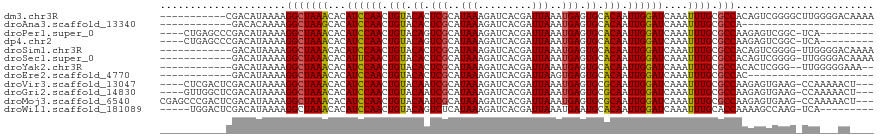
>dm3.chr3R 10316091 108 + 27905053 -----------CGACAUAAAAGGCUAAACACAUCCAACUGUACACUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCACAGUCGGGGCUUGGGGACAAAA -----------((((......(((((((...((((((.(((.((((((..(((.........)))..)))))).))).))))))....)))).)))...))))....(((....))).. ( -26.40, z-score = -2.14, R) >droAna3.scaffold_13340 481269 85 + 23697760 ------------GACACAAAAGGCUAAGCACAUCCAACUGUACACUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCA---------------------- ------------.........(((((((...((((((.(((.((((((..(((.........)))..)))))).))).))))))....)))).))).---------------------- ( -20.40, z-score = -2.74, R) >droPer1.super_0 7799613 105 - 11822988 ----CUGAGCCCGACAUAAAAGGCUAAACACAUCCAACUGUACAGUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCAAGAGUCGGC-UCA--------- ----..(((((.(((......(((((((...((((((.(((.((.(((..(((.........)))..))).)).))).))))))....)))).)))....))))))-)).--------- ( -27.50, z-score = -2.40, R) >dp4.chr2 16374924 105 + 30794189 ----CUGAGCCCGACAUAAAAGGCUAAACACAUCCAACUGUACAGUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCAAGAGUCGGC-UCA--------- ----..(((((.(((......(((((((...((((((.(((.((.(((..(((.........)))..))).)).))).))))))....)))).)))....))))))-)).--------- ( -27.50, z-score = -2.40, R) >droSim1.chr3R 16376764 106 - 27517382 ------------GACAUAAAAGGCUAAACACAUCCAACUGUACACUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCACAGUCGGGG-UUGGGGACAAAA ------------(((......(((((((...((((((.(((.((((((..(((.........)))..)))))).))).))))))....)))).)))...)))....-(((....))).. ( -24.50, z-score = -1.68, R) >droSec1.super_0 9394833 106 - 21120651 ------------GACAUAAAAGGCUAAACACAUUCAACUGUACACUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCACAGUCGGGG-UUGGGGACAAAA ------------(((......(((((((...((((((.(((.((((((..(((.........)))..)))))).))).))))))....)))).)))...)))....-(((....))).. ( -21.60, z-score = -0.99, R) >droYak2.chr3R 12710008 103 - 28832112 ------------GACAUAAAAGGCUAAACACAUCCAACUGUACACUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCACACUCGGG--UUGGGGGAAA-- ------------.........(((((((...((((((.(((.((((((..(((.........)))..)))))).))).))))))....)))).)))...(((...--...)))....-- ( -22.30, z-score = -1.28, R) >droEre2.scaffold_4770 9847311 86 + 17746568 ------------GACAUAAAAGGCUAAACACAUCCAACUGUACACUCGCAUAAAGAUCACGAUUAAGUGAGUGCACAAUUGGAUCAAAUUUGCGCCAC--------------------- ------------.........(((((((...((((((.(((.(((((((.(((.........))).))))))).))).))))))....)))).)))..--------------------- ( -24.60, z-score = -4.50, R) >droVir3.scaffold_13047 4622099 111 - 19223366 ----CUCGACUCGACAUAAAAGGCUAAACACAUCCAACUGUACAAUCGCAUAAAGAUCACGAUUAAAUGAGUGCGCAAUUGGAUCAAAUUUGCGCCAAGAGUGAAG-CCAAAAACU--- ----.................((((...(((............(((((...........)))))......(.((((((...........)))))))....))).))-)).......--- ( -18.80, z-score = -0.16, R) >droGri2.scaffold_14830 5373062 111 + 6267026 ----GUUGGCUCGACAUAAAAGGCUAAACACAUCCAACUGUACAAUCGCAUAAAGAUCACGAUUAAAUGAGUGCGCAAUUGGAUCAAAUUUGCGCCAAGAGUGAAG-CCAAAAACU--- ----.((((((...(((....(((((((...((((((.((..((....(((...(((....)))..)))..))..)).))))))....)))).)))....))).))-)))).....--- ( -23.30, z-score = -0.93, R) >droMoj3.scaffold_6540 16100663 115 - 34148556 CGAGCCCGACUCGACAUAAAAGGCUAAACACAUCCAACUGUACAAUCGCAUAAAGAUCACGAUUAAAUGAGUGCGCAAUUGGAUCAAAUUUGCGCCAAGAGUGAAG-CCAAAAACU--- ((((.....))))........((((...(((............(((((...........)))))......(.((((((...........)))))))....))).))-)).......--- ( -22.30, z-score = -0.89, R) >droWil1.scaffold_181089 2397643 104 - 12369635 -----UGGACUCGACAUAAAAGGCUAAACACAUCCAACUGUACAGUCUCAUAAAGAUCACGAUUAAAUGAAUGCACAAUUGGAUCAAAUUUGCACCAAAAGCCAAG-UCA--------- -----..((((...........((.......((((((.(((.((...((((...(((....)))..)))).)).))).)))))).......))...........))-)).--------- ( -16.29, z-score = -0.49, R) >consensus ___________CGACAUAAAAGGCUAAACACAUCCAACUGUACACUCGCAUAAAGAUCACGAUUAAAUGAGUGCACAAUUGGAUCAAAUUUGCGCCAAAAGCGGGG_UCA___AC____ .....................(((((((...((((((.(((.((.(((..(((.........)))..))).)).))).))))))....)))).)))....................... (-14.42 = -14.25 + -0.17)

| Location | 10,316,091 – 10,316,199 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Shannon entropy | 0.39403 |
| G+C content | 0.41740 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -18.81 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10316091 108 - 27905053 UUUUGUCCCCAAGCCCCGACUGUGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGAGUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCG----------- ................((((...(((..((((...((((((((((((((((...(((.........)))...))))))))))))))))..)))).)))......))))----------- ( -33.60, z-score = -4.22, R) >droAna3.scaffold_13340 481269 85 - 23697760 ----------------------UGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGAGUGUACAGUUGGAUGUGCUUAGCCUUUUGUGUC------------ ----------------------.((((((((....((((((((((((((((...(((.........)))...))))))))))))))))..((...))..))))))))------------ ( -29.90, z-score = -4.13, R) >droPer1.super_0 7799613 105 + 11822988 ---------UGA-GCCGACUCUUGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGACUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCGGGCUCAG---- ---------(((-((((((....(((..((((...(((((((((((((.((...(((.........)))...)).)))))))))))))..)))).)))......))).)))))).---- ( -37.50, z-score = -4.20, R) >dp4.chr2 16374924 105 - 30794189 ---------UGA-GCCGACUCUUGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGACUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCGGGCUCAG---- ---------(((-((((((....(((..((((...(((((((((((((.((...(((.........)))...)).)))))))))))))..)))).)))......))).)))))).---- ( -37.50, z-score = -4.20, R) >droSim1.chr3R 16376764 106 + 27517382 UUUUGUCCCCAA-CCCCGACUGUGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGAGUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUC------------ ............-....(((...(((..((((...((((((((((((((((...(((.........)))...))))))))))))))))..)))).)))......)))------------ ( -31.20, z-score = -3.80, R) >droSec1.super_0 9394833 106 + 21120651 UUUUGUCCCCAA-CCCCGACUGUGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGAGUGUACAGUUGAAUGUGUUUAGCCUUUUAUGUC------------ ............-....(((...(((..((((...((.(((((((((((((...(((.........)))...))))))))))))).))..)))).)))......)))------------ ( -24.40, z-score = -2.02, R) >droYak2.chr3R 12710008 103 + 28832112 --UUUCCCCCAA--CCCGAGUGUGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGAGUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUC------------ --..........--.........(((..((((...((((((((((((((((...(((.........)))...))))))))))))))))..)))).))).........------------ ( -30.00, z-score = -3.56, R) >droEre2.scaffold_4770 9847311 86 - 17746568 ---------------------GUGGCGCAAAUUUGAUCCAAUUGUGCACUCACUUAAUCGUGAUCUUUAUGCGAGUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUC------------ ---------------------..(((..((((...((((((((((((((((.(.(((.........))).).))))))))))))))))..)))).))).........------------ ( -31.30, z-score = -4.78, R) >droVir3.scaffold_13047 4622099 111 + 19223366 ---AGUUUUUGG-CUUCACUCUUGGCGCAAAUUUGAUCCAAUUGCGCACUCAUUUAAUCGUGAUCUUUAUGCGAUUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCGAGUCGAG---- ---.....((((-(((.((....(((..((((...(((((((((..((........(((((.........)))))))..)))))))))..)))).)))......)).))))))).---- ( -29.40, z-score = -1.66, R) >droGri2.scaffold_14830 5373062 111 - 6267026 ---AGUUUUUGG-CUUCACUCUUGGCGCAAAUUUGAUCCAAUUGCGCACUCAUUUAAUCGUGAUCUUUAUGCGAUUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCGAGCCAAC---- ---.....((((-(((.((....(((..((((...(((((((((..((........(((((.........)))))))..)))))))))..)))).)))......)).))))))).---- ( -32.10, z-score = -2.77, R) >droMoj3.scaffold_6540 16100663 115 + 34148556 ---AGUUUUUGG-CUUCACUCUUGGCGCAAAUUUGAUCCAAUUGCGCACUCAUUUAAUCGUGAUCUUUAUGCGAUUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCGAGUCGGGCUCG ---.(..(((((-(((.((....(((..((((...(((((((((..((........(((((.........)))))))..)))))))))..)))).)))......)).))))))))..). ( -29.70, z-score = -1.07, R) >droWil1.scaffold_181089 2397643 104 + 12369635 ---------UGA-CUUGGCUUUUGGUGCAAAUUUGAUCCAAUUGUGCAUUCAUUUAAUCGUGAUCUUUAUGAGACUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCGAGUCCA----- ---------.((-((((((....(((..((((...(((((((((((((.((......((((((...)))))))).)))))))))))))..)))).)))......))))))))..----- ( -34.80, z-score = -4.54, R) >consensus ____GU___UGA_CCCCACUCUUGGCGCAAAUUUGAUCCAAUUGUGCACUCAUUUAAUCGUGAUCUUUAUGCGAGUGUACAGUUGGAUGUGUUUAGCCUUUUAUGUCG___________ .......................(((..((((...(((((((((.(((.((...(((.........)))...)).))).)))))))))..)))).)))..................... (-18.81 = -18.90 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:11 2011