
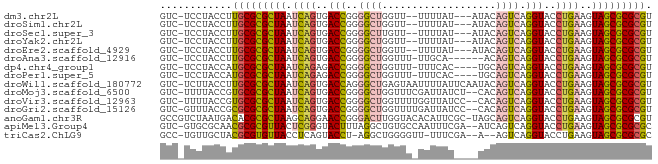
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,425,800 – 7,425,876 |
| Length | 76 |
| Max. P | 0.999527 |

| Location | 7,425,800 – 7,425,876 |
|---|---|
| Length | 76 |
| Sequences | 15 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Shannon entropy | 0.49212 |
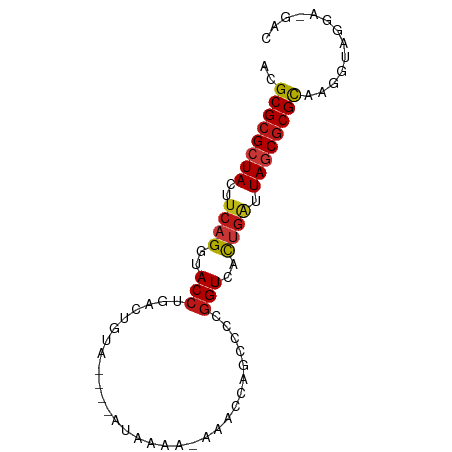
| G+C content | 0.54450 |
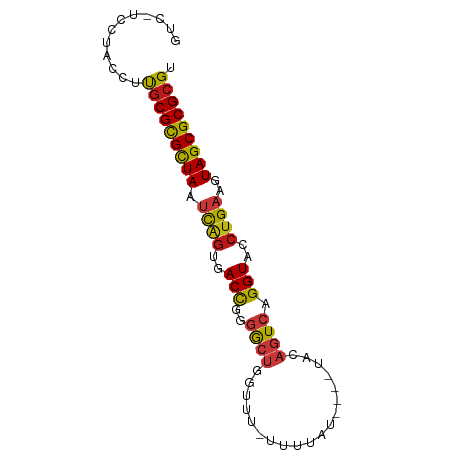
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7425800 76 + 23011544 GUC-UCCUACCUUGCGCGCUAAUCAGUGACCGGGGCUGGUU--UUUUAU---AUACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((...--......---...))))).)))..))))..)))))))).. ( -31.70, z-score = -3.54, R) >droSim1.chr2L 7217370 76 + 22036055 GUC-UCCUACCUUGCGCGCUAAUCAGUGACCGGGGCUGGUU--UUUUAU---AUACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((...--......---...))))).)))..))))..)))))))).. ( -31.70, z-score = -3.54, R) >droSec1.super_3 2941260 76 + 7220098 GUC-UCCUACCUUGCGCGCUAAUCAGUGACCGGGGCUUGUU--UUUUAU---AUACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..((((.((.--......---..)))))).)))..))))..)))))))).. ( -28.80, z-score = -3.03, R) >droYak2.chr2L 16850578 76 - 22324452 GUC-UCCUACCUUGCGCGCUAAUCAGUGACCGGGGCUGGUU--UUUUAU---AUACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((...--......---...))))).)))..))))..)))))))).. ( -31.70, z-score = -3.54, R) >droEre2.scaffold_4929 16344526 76 + 26641161 GUC-UCCUACCUUGCGCGCUAAUCAGUGACCGGGGCUGGUU--UUUUAU---AUACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((...--......---...))))).)))..))))..)))))))).. ( -31.70, z-score = -3.54, R) >droAna3.scaffold_12916 15333995 74 + 16180835 GUC-UCCUACCUUGCGCGCUAAUCAGUGACCGGGGCUGGUUU-UUGCA------ACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((....-.....------.))))).)))..))))..)))))))).. ( -31.90, z-score = -3.03, R) >dp4.chr4_group1 4175740 76 + 5278887 GUC-UCCUACCAUGCGCGCUAAUCAGAGACCGGGGCUGGUUU-UUUCAC----UGCAGUCAGGUACCUGAAGUAGCGCGCGU ...-.......((((((((((.((((..(((..(((((....-......----..))))).)))..))))..)))))))))) ( -31.70, z-score = -2.94, R) >droPer1.super_5 5727157 76 - 6813705 GUC-UCCUACCAUGCGCGCUAAUCAGAGACCGGGGCUGGUUU-UUUCAC----UGCAGUCAGGUACCUGAAGUAGCGCGCGU ...-.......((((((((((.((((..(((..(((((....-......----..))))).)))..))))..)))))))))) ( -31.70, z-score = -2.94, R) >droWil1.scaffold_180772 3648555 81 - 8906247 GUC-UCUUACCUUGCGCGCUAAUCAGUGACCAGGGCUGAGUAAUUUUAUUCAAUACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((.((...........)).))))).)))..))))..)))))))).. ( -29.00, z-score = -3.28, R) >droMoj3.scaffold_6500 13952913 79 - 32352404 GUC-UUUUACCGUGCGCGCUAAUCAGUGACCGGGGCUGGUUUCGAUUAUCU--CACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((.............--..))))).)))..))))..)))))))).. ( -31.46, z-score = -2.90, R) >droVir3.scaffold_12963 4786196 79 - 20206255 GUC-UUUUACCGUGCGCGCUAAUCAGUGACCGGGGCUGGUUUUGGUUAUCC--CACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.........((((((((.((((..(((..(((((....(((.....)--))))))).)))..))))..)))))))).. ( -32.70, z-score = -2.90, R) >droGri2.scaffold_15126 5008400 79 + 8399593 GUC-GUUUACCGCGCGCGCUAAUCAGUGACCGGGGCUGGUUUUGAUUAUCC--CACAGUCAGGUACCUGAAGUAGCGCGCGU ...-.......((((((((((.((((..(((..(((((.............--..))))).)))..))))..)))))))))) ( -34.46, z-score = -3.31, R) >anoGam1.chr3R 39066347 81 + 53272125 GCCGUCUAAUGACACGCGCUAAGCAGGAACCGGGACUUGGUACACAUUCGC-UAGCAGUCAGGUACCUGAAGUAGCGCGCGU ((.(((....)))..((((((..((((.(((..((((((((........))-))..)))).))).))))...)))))))).. ( -33.10, z-score = -2.82, R) >apiMel3.Group4 8490912 79 - 10796202 GUC-GUGCGCAACGCGCGUUACUCGGGUACUUUAGGCUGUGCCAAUUUCGA--AUCAGUCAGGUACCUGAAGUAGCGCGCGC ...-........((((((((((((((((((((..(((((...(......).--..)))))))))))))).))))))))))). ( -37.10, z-score = -4.00, R) >triCas2.ChLG9 5914742 75 - 15222296 GCC-UGUUGCUACGCGUGUUACCUCAGUACCU-AGGCUGGGGUU-UUUCGA--A--AGUCAGGUACCUGAAGUAGCGCGCGC ...-........((((((((((.(((((((((-.((((..((..-..))..--.--))))))))).)))).)))))))))). ( -31.90, z-score = -2.83, R) >consensus GUC_UCCUACCUUGCGCGCUAAUCAGUGACCGGGGCUGGUUU_UUUUAU____UACAGUCAGGUACCUGAAGUAGCGCGCGU ............(((((((((.((((..(((..((((...................)))).)))..))))..))))))))). (-24.06 = -24.18 + 0.12)
| Location | 7,425,800 – 7,425,876 |
|---|---|
| Length | 76 |
| Sequences | 15 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Shannon entropy | 0.49212 |
| G+C content | 0.54450 |
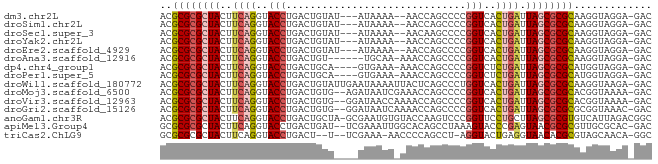
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.51 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7425800 76 - 23011544 ACGCGCGCUACUUCAGGUACCUGACUGUAU---AUAAAA--AACCAGCCCCGGUCACUGAUUAGCGCGCAAGGUAGGA-GAC ..((((((((..((((..(((.(.(((...---......--...)))..).)))..)))).)))))))).........-... ( -25.30, z-score = -2.34, R) >droSim1.chr2L 7217370 76 - 22036055 ACGCGCGCUACUUCAGGUACCUGACUGUAU---AUAAAA--AACCAGCCCCGGUCACUGAUUAGCGCGCAAGGUAGGA-GAC ..((((((((..((((..(((.(.(((...---......--...)))..).)))..)))).)))))))).........-... ( -25.30, z-score = -2.34, R) >droSec1.super_3 2941260 76 - 7220098 ACGCGCGCUACUUCAGGUACCUGACUGUAU---AUAAAA--AACAAGCCCCGGUCACUGAUUAGCGCGCAAGGUAGGA-GAC ..((((((((..((((..(((.(..(((..---......--.)))..)...)))..)))).)))))))).........-... ( -24.00, z-score = -2.04, R) >droYak2.chr2L 16850578 76 + 22324452 ACGCGCGCUACUUCAGGUACCUGACUGUAU---AUAAAA--AACCAGCCCCGGUCACUGAUUAGCGCGCAAGGUAGGA-GAC ..((((((((..((((..(((.(.(((...---......--...)))..).)))..)))).)))))))).........-... ( -25.30, z-score = -2.34, R) >droEre2.scaffold_4929 16344526 76 - 26641161 ACGCGCGCUACUUCAGGUACCUGACUGUAU---AUAAAA--AACCAGCCCCGGUCACUGAUUAGCGCGCAAGGUAGGA-GAC ..((((((((..((((..(((.(.(((...---......--...)))..).)))..)))).)))))))).........-... ( -25.30, z-score = -2.34, R) >droAna3.scaffold_12916 15333995 74 - 16180835 ACGCGCGCUACUUCAGGUACCUGACUGU------UGCAA-AAACCAGCCCCGGUCACUGAUUAGCGCGCAAGGUAGGA-GAC ..((((((((..((((.....((((((.------.((..-......))..)))))))))).)))))))).........-... ( -27.50, z-score = -2.32, R) >dp4.chr4_group1 4175740 76 - 5278887 ACGCGCGCUACUUCAGGUACCUGACUGCA----GUGAAA-AAACCAGCCCCGGUCUCUGAUUAGCGCGCAUGGUAGGA-GAC ..((((((((..((((..(((.(.(((..----.(....-)...)))..).)))..)))).)))))))).........-... ( -27.40, z-score = -2.06, R) >droPer1.super_5 5727157 76 + 6813705 ACGCGCGCUACUUCAGGUACCUGACUGCA----GUGAAA-AAACCAGCCCCGGUCUCUGAUUAGCGCGCAUGGUAGGA-GAC ..((((((((..((((..(((.(.(((..----.(....-)...)))..).)))..)))).)))))))).........-... ( -27.40, z-score = -2.06, R) >droWil1.scaffold_180772 3648555 81 + 8906247 ACGCGCGCUACUUCAGGUACCUGACUGUAUUGAAUAAAAUUACUCAGCCCUGGUCACUGAUUAGCGCGCAAGGUAAGA-GAC ..((((((((..((((..(((.(.(((.(((......)))....))).)..)))..)))).)))))))).........-... ( -26.80, z-score = -2.91, R) >droMoj3.scaffold_6500 13952913 79 + 32352404 ACGCGCGCUACUUCAGGUACCUGACUGUG--AGAUAAUCGAAACCAGCCCCGGUCACUGAUUAGCGCGCACGGUAAAA-GAC ..((((((((..((((..(((.(.(((((--(.....)))....)))..).)))..)))).)))))))).........-... ( -26.20, z-score = -2.39, R) >droVir3.scaffold_12963 4786196 79 + 20206255 ACGCGCGCUACUUCAGGUACCUGACUGUG--GGAUAACCAAAACCAGCCCCGGUCACUGAUUAGCGCGCACGGUAAAA-GAC ..((((((((..((((.....((((((.(--((..............))))))))))))).)))))))).........-... ( -30.44, z-score = -3.37, R) >droGri2.scaffold_15126 5008400 79 - 8399593 ACGCGCGCUACUUCAGGUACCUGACUGUG--GGAUAAUCAAAACCAGCCCCGGUCACUGAUUAGCGCGCGCGGUAAAC-GAC .(((((((((..((((.....((((((.(--((..............))))))))))))).)))))))))((.....)-).. ( -32.24, z-score = -3.42, R) >anoGam1.chr3R 39066347 81 - 53272125 ACGCGCGCUACUUCAGGUACCUGACUGCUA-GCGAAUGUGUACCAAGUCCCGGUUCCUGCUUAGCGCGUGUCAUUAGACGGC ((((((((((...((((.(((.((((....-(((....)))....))))..))).))))..))))))))))........... ( -30.10, z-score = -1.87, R) >apiMel3.Group4 8490912 79 + 10796202 GCGCGCGCUACUUCAGGUACCUGACUGAU--UCGAAAUUGGCACAGCCUAAAGUACCCGAGUAACGCGCGUUGCGCAC-GAC (((((((.(((((..(((((.(((.....--))).....(((...)))....))))).))))).))))))).......-... ( -25.30, z-score = -1.10, R) >triCas2.ChLG9 5914742 75 + 15222296 GCGCGCGCUACUUCAGGUACCUGACU--U--UCGAAA-AACCCCAGCCU-AGGUACUGAGGUAACACGCGUAGCAACA-GGC (((((.(.((((((((.((((((...--.--......-..........)-))))))))))))).).))))).......-... ( -24.55, z-score = -1.67, R) >consensus ACGCGCGCUACUUCAGGUACCUGACUGUA____AUAAAA_AAACCAGCCCCGGUCACUGAUUAGCGCGCAAGGUAGGA_GAC ..((((((((..((((..(((..............................)))..)))).))))))))............. (-19.16 = -19.51 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:41 2011