
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,269,264 – 10,269,382 |
| Length | 118 |
| Max. P | 0.558721 |

| Location | 10,269,264 – 10,269,382 |
|---|---|
| Length | 118 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.25 |
| Shannon entropy | 0.54305 |
| G+C content | 0.31787 |
| Mean single sequence MFE | -19.81 |
| Consensus MFE | -9.50 |
| Energy contribution | -9.28 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
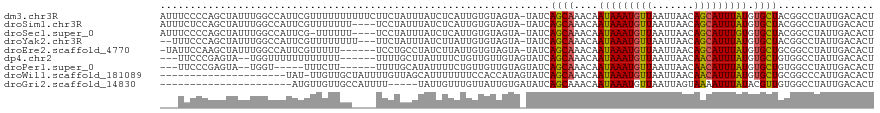
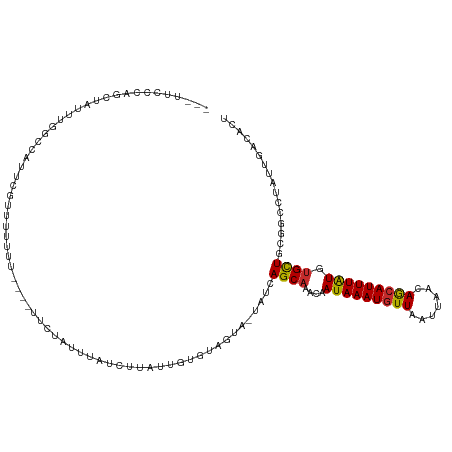
>dm3.chr3R 10269264 118 - 27905053 AUUUCCCCAGCUAUUUGGCCAUUCGUUUUUUUUUUCUUCUAUUUAUCUCAUUGUGUAGUA-UAUCAGCAAACAAUAAAUGUUAAUUAACAGCAUUUAUGUGCUACGGCCUAUUGACACU ................((((..................((((.((......)).))))..-....((((....(((((((((.......))))))))).))))..)))).......... ( -19.60, z-score = -1.48, R) >droSim1.chr3R 16329331 114 + 27517382 AUUUCUCCAGCUAUUUGGCCAUUCGUUUUUUU----UCCUAUUUAUCUCAUUGUGUAGUA-UAUCAGCAAACAAUAAAUGUUAAUUAACAGCAUUUAUGUGCUACGGCCUAUUGACACU ................((((............----..((((.((......)).))))..-....((((....(((((((((.......))))))))).))))..)))).......... ( -19.80, z-score = -1.41, R) >droSec1.super_0 9348720 113 + 21120651 AUUUCCCCAGCUAUUUGGCCAUUCG-UUUUUU----UCCUAUUUAUCUCAUUGUGUAGUA-UAUCAGCAAACAAUAAAUGUUAAUUAACAGCAUUUGUGUGCUACGGCCUAUUGACACU ................((((.....-......----..((((.((......)).))))..-....((((....(((((((((.......))))))))).))))..)))).......... ( -19.80, z-score = -1.06, R) >droYak2.chr3R 12661718 113 + 28832112 --UUUCCCAGCUAUUUGGCCAUUCGUUUUUUUU---UUCUAUUUAUCUUAUUGUGUAGUA-UAUCAGCAAACAAUAAAUGUUAAUUAACAGCAUUUAUGUGCUACGGCCUAUUGACACU --..............((((.............---..((((.((......)).))))..-....((((....(((((((((.......))))))))).))))..)))).......... ( -19.60, z-score = -1.25, R) >droEre2.scaffold_4770 9801950 111 - 17746568 -UAUUCCAAGCUAUUUGGCCAUUCGUUUUU------UCCUGCCUAUCUUAUUGUGUAGUA-UAUCAGCAAACAAUAAAUGUUAAUUAACAGCAUUUAUGUGCUGCGGCCUAUUGACACU -...............((((..........------..((((.((......)).))))..-...(((((....(((((((((.......))))))))).))))).)))).......... ( -23.30, z-score = -1.31, R) >dp4.chr2 16322793 108 - 30794189 ---UUCCCGAGUA--UGGUUUUUUUUUUUU------UUUUGCUUAUUUUCUGUUGUUGUAGUAUCAGCAAACAAUAAAUGUUAAUUAACAACAUUUAUGUGCUGUGGCCUAUUGACACU ---.....(((((--...............------...)))))......((((...((((...(((((....(((((((((.......))))))))).)))))....)))).)))).. ( -18.57, z-score = -0.62, R) >droPer1.super_0 7746833 103 + 11822988 ---UUCCCGAGUA--UGGU-----UUUCUU------UUUUGCAUAUUUUCUGUUGUUGUAGUAUCAGCAAACAAUAAAUGUUAAUUAACAACAUUUAUGUGCUGUGGCCUAUUGACACU ---.....(((((--((..-----......------.....)))))))..((((...((((...(((((....(((((((((.......))))))))).)))))....)))).)))).. ( -19.52, z-score = -0.67, R) >droWil1.scaffold_181089 2345517 97 + 12369635 ---------------------UAU-UUGUUGCUAUUUUGUUAGCAUUUUUUUCCACCAUAGUAUCAGCAAACAAUAAAUGUUAAUUAACAACAUUUAUGUGCUGCGGCCCAUUGACACU ---------------------...-.((((((((......))))................((..(((((....(((((((((.......))))))))).)))))..)).....)))).. ( -19.50, z-score = -1.68, R) >droGri2.scaffold_14830 5315966 92 - 6267026 ----------------------AUGUUGUUGCCAUUUU-----UAUUGUUUGUUAUUGUGAUAUCAGCAAACAAUAAAUGUUAAUUAGUAAAAUUUAUACGUUGUGGCCUAUUGACACU ----------------------.(((((..(((((...-----(((((((((((((......)).)))))))))))(((((.((((.....))))...))))))))))....))))).. ( -18.60, z-score = -1.29, R) >consensus ___UUCCCAGCUAUUUGGCCAUUCGUUUUUUU____UUCUAUUUAUCUUAUUGUGUAGUA_UAUCAGCAAACAAUAAAUGUUAAUUAACAGCAUUUAUGUGCUGCGGCCUAUUGACACU .................................................................((((....(((((((((.......))))))))).))))................ ( -9.50 = -9.28 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:05 2011