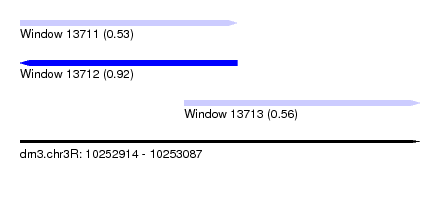
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,252,914 – 10,253,087 |
| Length | 173 |
| Max. P | 0.920655 |

| Location | 10,252,914 – 10,253,008 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Shannon entropy | 0.14386 |
| G+C content | 0.56599 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10252914 94 + 27905053 AACGGGCGCCGCCUGGCUUAAGAAUUUCUUAUCAGCUGUCAUGGUGUGGCUGCCUGGCGAUGUUGCUCCCGUUGCCGAUGAUGUGGCCGAUGUU ((((((.((((((.(((.((((.....))))..(((..(......)..)))))).)))).....)).))))))((((......))))....... ( -32.30, z-score = -0.60, R) >droSim1.chr3R 16312553 82 - 27517382 AACGGGCGCCGCCUGGCUUAAGAAUUUCUUAUCAGCUGUCAUGGUGUGGCUGCCUGGCGAUGUUGCUCCCGUUGCCGAUGUU------------ ...(((((((((((((((((((.....))))..)))).....)).))))).))))(((((((.......)))))))......------------ ( -28.70, z-score = -1.01, R) >droEre2.scaffold_4770 9785883 82 + 17746568 AACGGGCGCCGCCUGGCUUAAGAAUUUCUUAUCAGCUGUCAUGGUGUGGCUGCCUGGCGAUGUUGCACCCGUUGCCGAUG-UG----------- ((((((.((((((.(((.((((.....))))..(((..(......)..)))))).)))).....)).)))))).......-..----------- ( -28.90, z-score = -0.88, R) >consensus AACGGGCGCCGCCUGGCUUAAGAAUUUCUUAUCAGCUGUCAUGGUGUGGCUGCCUGGCGAUGUUGCUCCCGUUGCCGAUG_UG___________ ...(((((((((((((((((((.....))))..)))).....)).))))).))))(((((((.......))))))).................. (-28.70 = -28.70 + -0.00)

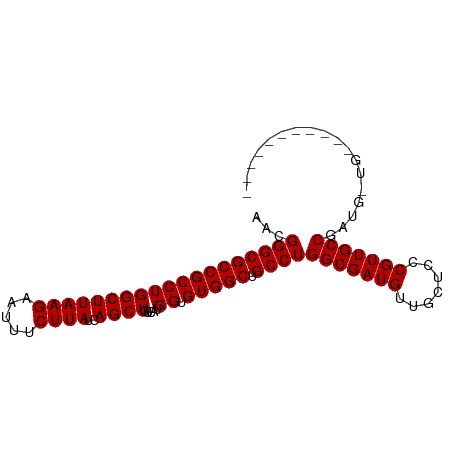
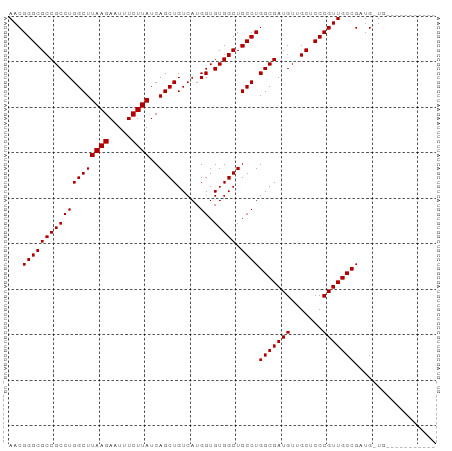
| Location | 10,252,914 – 10,253,008 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Shannon entropy | 0.14386 |
| G+C content | 0.56599 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -24.93 |
| Energy contribution | -24.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10252914 94 - 27905053 AACAUCGGCCACAUCAUCGGCAACGGGAGCAACAUCGCCAGGCAGCCACACCAUGACAGCUGAUAAGAAAUUCUUAAGCCAGGCGGCGCCCGUU .......(((........)))((((((.((.....((((.((((((............)))..((((.....)))).))).)))))).)))))) ( -29.40, z-score = -1.86, R) >droSim1.chr3R 16312553 82 + 27517382 ------------AACAUCGGCAACGGGAGCAACAUCGCCAGGCAGCCACACCAUGACAGCUGAUAAGAAAUUCUUAAGCCAGGCGGCGCCCGUU ------------.........((((((.((.....((((.((((((............)))..((((.....)))).))).)))))).)))))) ( -25.10, z-score = -1.51, R) >droEre2.scaffold_4770 9785883 82 - 17746568 -----------CA-CAUCGGCAACGGGUGCAACAUCGCCAGGCAGCCACACCAUGACAGCUGAUAAGAAAUUCUUAAGCCAGGCGGCGCCCGUU -----------..-.......(((((((((.....((((.((((((............)))..((((.....)))).))).))))))))))))) ( -28.90, z-score = -2.38, R) >consensus ___________CA_CAUCGGCAACGGGAGCAACAUCGCCAGGCAGCCACACCAUGACAGCUGAUAAGAAAUUCUUAAGCCAGGCGGCGCCCGUU .....................((((((.((.....((((.((((((............)))..((((.....)))).))).)))))).)))))) (-24.93 = -24.93 + 0.00)

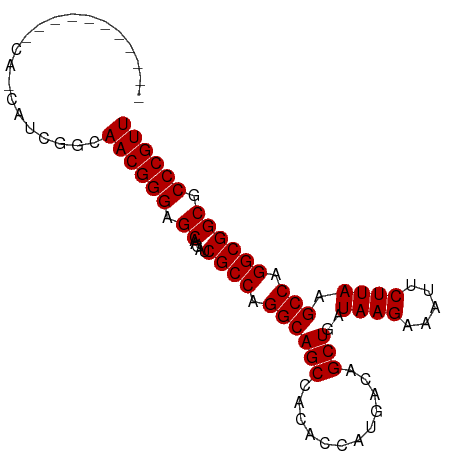
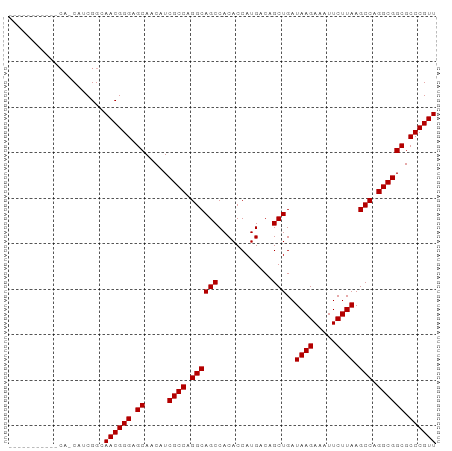
| Location | 10,252,985 – 10,253,087 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.43339 |
| G+C content | 0.56160 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -20.74 |
| Energy contribution | -23.17 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10252985 102 + 27905053 UUGCCGAUGAUGUGGCCGAUGUUGCCGAUGUUGCCAAUGUUGCUCCUGUUGCAAGUGCAGCGGCAACAUUGCUUCCGUAUUGGCGGGUUCGCGGCAUGGUUG ....((((.((((.((.((..(((((((((..(.((((((((((...(((((....)))))))))))))))...)..)))))))))..)))).)))).)))) ( -41.90, z-score = -1.64, R) >droSim1.chr3R 16312624 90 - 27517382 ------------UUGCCGAUGUUGCUCCUGUUGCCAAUGUUGCUCCUGUUGCAAGUGCAGCGGCAACAUUGCUUCCGUAUUGGCGGGUUCGCGGCAUGGUUG ------------..(((.(((((((.......((.(((((((((...(((((....)))))))))))))))).(((((....)))))...)))))))))).. ( -35.60, z-score = -1.66, R) >droEre2.scaffold_4770 9785954 81 + 17746568 ---------------------UUGCCGAUGUGGCCGAUGUUGCUCCUGUUGCAAGUGCAGCGGCAACAUUGCUUCCGUAUUGGCGAGUUCGCGGCAUGGUUG ---------------------((((((((((((.((((((((((...(((((....)))))))))))))))...))))))))))))................ ( -32.90, z-score = -1.66, R) >droWil1.scaffold_181089 2327642 78 - 12369635 ------------------------CUCCUGUUGCAGCCAUUGCCGAGGCAGCUACUACUAGUGCAGCAUUGCUUCCGUAUUGGCGUGUGCGAGGCAUGGUUG ------------------------.........(((((((.((((((((((((.(.......).)))..))))))(((((......))))).)))))))))) ( -27.20, z-score = -0.14, R) >consensus _____________________UUGCCCAUGUUGCCAAUGUUGCUCCUGUUGCAAGUGCAGCGGCAACAUUGCUUCCGUAUUGGCGGGUUCGCGGCAUGGUUG .......................(((.(((((((((((((((((.((((.......)))).))))))))))..(((((....)))))...)))))))))).. (-20.74 = -23.17 + 2.44)

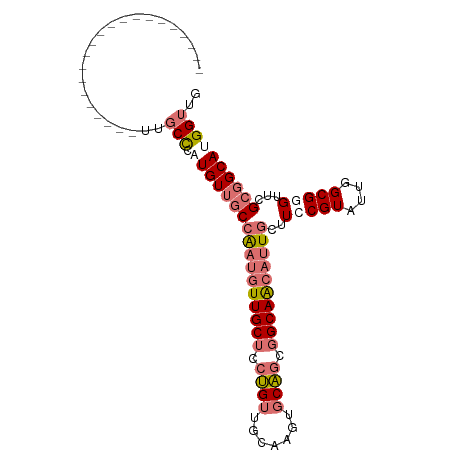
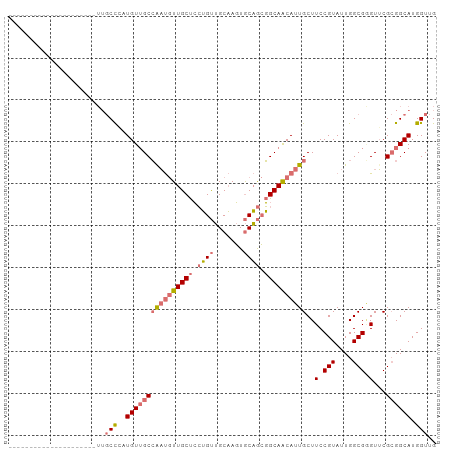
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:03 2011