
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,250,971 – 10,251,067 |
| Length | 96 |
| Max. P | 0.754157 |

| Location | 10,250,971 – 10,251,067 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Shannon entropy | 0.39460 |
| G+C content | 0.47352 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -17.97 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
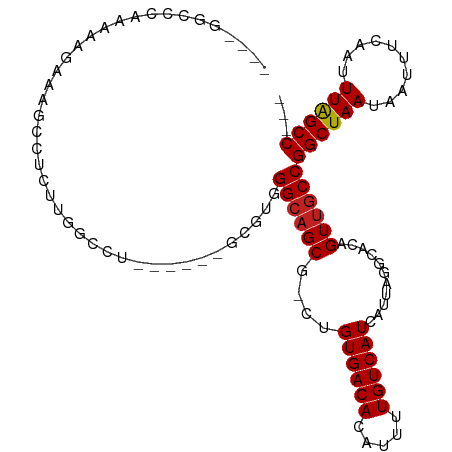
>dm3.chr3R 10250971 96 + 27905053 ----GGCCCAAAAAGAAAGCUUUUUGGCCU------GCGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUUAAUUUAGCC--- ----((((.((((((....)))))))))).------.....(((((((-((((((((.....)))))).....)))...))))))((((((..........))))))--- ( -32.50, z-score = -2.19, R) >droSim1.chr3R 16310614 96 - 27517382 ----GGCCCGAAAAGAAAGCUUUUUGGCCU------GCGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- ----((((.((((((....)))))))))).------.....(((((((-((((((((.....)))))).....)))...))))))((((((..........))))))--- ( -32.20, z-score = -1.92, R) >droSec1.super_0 9330682 96 - 21120651 ----GGCCCGAAAAGAAAGCUUUUUGGCCU------GCGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- ----((((.((((((....)))))))))).------.....(((((((-((((((((.....)))))).....)))...))))))((((((..........))))))--- ( -32.20, z-score = -1.92, R) >droYak2.chr3R 12642564 95 - 28832112 ----GGCCCGAAAAGAAAGG-UUUUGGCCU------GCGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- ----((((.(((........-))).)))).------.....(((((((-((((((((.....)))))).....)))...))))))((((((..........))))))--- ( -28.60, z-score = -0.85, R) >droEre2.scaffold_4770 9783995 96 + 17746568 ----GGCCCGAAAAGAAAGGCUUUUGGCCU------GCGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- ----..............(((..(((((((------((((.....)))-).((((((.....))))))....)))).)))..)))((((((..........))))))--- ( -30.50, z-score = -1.04, R) >droAna3.scaffold_13340 426232 95 + 23697760 --UAAGCCC--AAAGAAAGC-UCCCAACCU------GCGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUCGCCGGCUAAUAAUUUCAAUUUGGCC--- --...(((.--...((((((-(((((....------...)))).)))(-((((((((.....)))))).....(((......))))))......)))).....))).--- ( -26.30, z-score = -0.90, R) >dp4.chr2 16304054 106 + 30794189 UUCCGAAGAAAGCAGAGAGACCGGAGACCGAAGACAGUGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- (((((................)))))...((.(((((((((.(((...-.)))..)))))..)))).))....((((....))))((((((..........))))))--- ( -27.29, z-score = -0.95, R) >droPer1.super_0 7727999 106 - 11822988 UUCCGAAGAAAGCAGAGAGACCGGAGACCGAAGACAGUGUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- (((((................)))))...((.(((((((((.(((...-.)))..)))))..)))).))....((((....))))((((((..........))))))--- ( -27.29, z-score = -0.95, R) >droVir3.scaffold_13047 4552315 95 - 19223366 ----AGCAGCAAAAGAAAGCGACUGCCCC-------GUGUGGGCAGCA-CUGUGACACAUUUUGUCAUCAUUAGCCACAGUUACCGGCUAAUAAUUUCAAUUUAGCC--- ----....((.......((.(.((((((.-------....))))))).-))((((((.....)))))).(((((((.........)))))))............)).--- ( -25.60, z-score = -1.58, R) >droMoj3.scaffold_6540 16022989 95 - 34148556 ----AGCGGCAAAAGAAAGCGACUGCCUC-------GUGUGGGCAGCA-CUGUGACACAUUUUGUCAUCAUUAGCCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- ----...(((((......(((.((((((.-------....))))))).-..((((((.....)))))).....)).....)))))((((((..........))))))--- ( -29.20, z-score = -1.73, R) >droGri2.scaffold_14830 5297927 102 + 6267026 ----GCCACCAAAAGAAAGCGACUUCCCCCACUCUCGUGUGGGCAGCA-GUGUGACACAUUUUGUCAUCAUUAGCCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC--- ----..............((((((...(((((......)))))..(((-((((((((.....))))).)))).))...)))))).((((((..........))))))--- ( -29.40, z-score = -2.98, R) >droWil1.scaffold_181089 2326083 91 - 12369635 -------AAAAAAAAAAAACAACU------------GCGUGGGCAGCGGCUGUGACACAUUUUGUCAUCAUUAGCCACAGUUGCCGGCUAAUAAUUUCAAUUUGGCCAAC -------.................------------.....((((((((((((((((.....))))))....))))...))))))((((((..........))))))... ( -26.70, z-score = -2.42, R) >consensus ____GGCCCAAAAAGAAAGCCUCUUGGCCU______GCGUGGGCAGCG_CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCC___ .........................................((((((..((((((((.....))))))....)).....))))))((((((..........))))))... (-17.97 = -18.08 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:15:01 2011