
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,245,348 – 10,245,493 |
| Length | 145 |
| Max. P | 0.878097 |

| Location | 10,245,348 – 10,245,443 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Shannon entropy | 0.42875 |
| G+C content | 0.52045 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -12.94 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
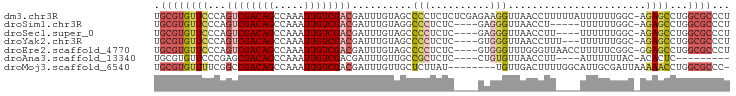
>dm3.chr3R 10245348 95 - 27905053 UGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUCUCGAGAAGGUUAACCUUUUUAUUUUUUGGC-AGAGCCUGGCGCCCU .((((((((...((((((((.....))))))))........(((........(((((((....))))))).......)))-.))))...))))... ( -28.26, z-score = -2.20, R) >droSim1.chr3R 16305070 86 + 27517382 UGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGGCCCUCUC----GAGGGUUAACCU-----UUUUUUGGC-AGAGCCUGGCGCCCU .(((((((((((((((((((.....)))))))).....((..(((((...----.)))))..))..-----.....))).-.))))...))))... ( -28.90, z-score = -1.60, R) >droSec1.super_0 9325137 87 + 21120651 UGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC----GAGGGUUAACCUU----UUUUUUGGC-AGAGCCUGGCGCCCU .(((((((((((((((((((.....))))))))......(((((((....----).)))))).....----.....))).-.))))...))))... ( -26.90, z-score = -1.51, R) >droYak2.chr3R 12636943 88 + 28832112 UGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC----GUGGGUUAACCUUU---UUUUUUGGC-AGAGCCUGGCGCCCU .(((((((((((((((((((.....))))))))......(((((((....----).))))))......---.....))).-.))))...))))... ( -26.90, z-score = -1.63, R) >droEre2.scaffold_4770 9778592 91 - 17746568 UGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC----GUGGGUUUGGGUUAACCUUUUUCGGC-GGAGCCUGGCGCCCU .((((((((((.((((((((.....))))))))((....(((((((((..----..)))...)))))).......)).).-)))))...))))... ( -30.00, z-score = -0.60, R) >droAna3.scaffold_13340 421413 78 - 23697760 UGCGUGUUCCCGAGCGACAGCCAAAUUGUCGACGAUUUGUUGCCGCUCUC----CUGUGUUAACCUU----AUUUUUUAC-ACACUC--------- ...((((....(((((.((((.((((((....)))))))))).)))))..----..(((..((....----..))..)))-))))..--------- ( -19.90, z-score = -2.84, R) >droMoj3.scaffold_6540 16015680 87 + 34148556 UGCGUGUUUUCGGCCGACAGCCAAAUUGUCGACGAUUUGUUGCUCUUAU--------UGUUGACUUUUGGCAUUGCGAUUAAAAACCUGGCGCCC- .(((((((((..(.((.((((((((..(((((((((..(.....)..))--------))))))).))))))..)))).)..)))))...))))..- ( -28.00, z-score = -2.68, R) >consensus UGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC____GAGGGUUAACCUU____UUUUUUGGC_AGAGCCUGGCGCCCU .((((((((...((((((((.....))))))))..........(((..........))).......................))))...))))... (-12.94 = -13.90 + 0.96)

| Location | 10,245,371 – 10,245,476 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.05 |
| Shannon entropy | 0.30199 |
| G+C content | 0.43274 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -16.97 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.755063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10245371 105 + 27905053 AUAAAAAGGUUAACCUU--CUCGAGAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUACC------ .......(((...((((--((....))))))(((.......(((((((.......)))))))......(((((....(((.....))))))))........))))))------ ( -27.90, z-score = -2.05, R) >droSim1.chr3R 16305090 99 - 27517382 --AAAAAGGUUAACCCU--C----GAGAGGGCCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUACC------ --.....((((..((((--.----...))))((((......(((((((.......)))))))))))..(((((....(((.....))))))))....))))......------ ( -25.70, z-score = -1.39, R) >droSec1.super_0 9325158 99 - 21120651 --AAAAAGGUUAACCCU--C----GAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUACC------ --.....(((...((((--.----...))))(((.......(((((((.......)))))))......(((((....(((.....))))))))........))))))------ ( -26.10, z-score = -1.54, R) >droYak2.chr3R 12636965 99 - 28832112 --AAAAAGGUUAACCCA------CGAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUACC------ --.....((((..((((------.((...(.....)...))(((((((.......)))))))))))..(((((....(((.....))))))))....))))......------ ( -26.10, z-score = -1.67, R) >droEre2.scaffold_4770 9778611 105 + 17746568 --AAAAAGGUUAACCCAAACCCACGAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUACC------ --.....((((..((((..(((.(....)))).........(((((((.......)))))))))))..(((((....(((.....))))))))....))))......------ ( -31.30, z-score = -2.67, R) >droVir3.scaffold_13047 4545951 94 - 19223366 --UAAAAAGUCAACA----------AUAAGAGCAACAAAUUGUCGACAAUUUGGCUGUCAGCCAAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUUC------- --......(((.(((----------((...........))))).)))..((((((.....))))))..(((((....(((.....)))))))).............------- ( -21.10, z-score = -3.04, R) >droGri2.scaffold_14830 5291421 94 + 6267026 --UGAAAAGUUAACA----------AUAUGAGCAACAAAUCGUCGACAAUUUGGCUGUCGACCAAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUUC------- --.............----------....((((........(((((((.......)))))))......(((((....(((.....)))))))).........))))------- ( -20.40, z-score = -2.06, R) >droAna3.scaffold_13340 421425 105 + 23697760 --AAUAAGGUUAACACA------GGAGAGCGGCAACAAAUCGUCGACAAUUUGGCUGUCGCUCGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAGUCAGUACGAGCUCC --.....((((...((.------...((((((((.(((((.(....).)))))..)))))))).....(((((....(((.....)))))))).........))...)))).. ( -27.60, z-score = -1.43, R) >droMoj3.scaffold_6540 16015705 94 - 34148556 --CCAAAAGUCAACA----------AUAAGAGCAACAAAUCGUCGACAAUUUGGCUGUCGGCCGAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUGC------- --.............----------......(((.....(((((((((.......)))))).)))...(((((....(((.....))))))))..........)))------- ( -19.10, z-score = -0.89, R) >consensus __AAAAAGGUUAACCC________GAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUACC______ .................................(((.....(((((((.......)))))))......(((((....(((.....)))))))).........)))........ (-16.97 = -16.92 + -0.05)



| Location | 10,245,371 – 10,245,476 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Shannon entropy | 0.30199 |
| G+C content | 0.43274 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -14.45 |
| Energy contribution | -14.34 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
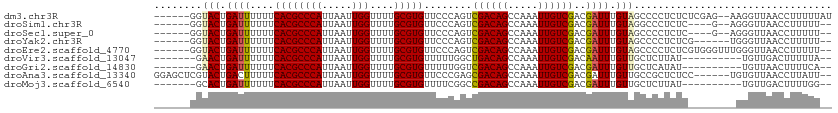
>dm3.chr3R 10245371 105 - 27905053 ------GGUACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUCUCGAG--AAGGUUAACCUUUUUAU ------(((((.((((....((((((((.....)))....)))))......((((((((.....)))))))))))).)).)))........(((--((((....))))))).. ( -29.00, z-score = -3.18, R) >droSim1.chr3R 16305090 99 + 27517382 ------GGUACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGGCCCUCUC----G--AGGGUUAACCUUUUU-- ------(((((.((((....((((((((.....)))....)))))......((((((((.....)))))))))))).)).((((((...----.--)))))).))).....-- ( -29.70, z-score = -2.71, R) >droSec1.super_0 9325158 99 + 21120651 ------GGUACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC----G--AGGGUUAACCUUUUU-- ------(((...........((((((((.....)))....)))))......((((((((.....))))))))......(((((((....----)--.))))))))).....-- ( -27.80, z-score = -2.64, R) >droYak2.chr3R 12636965 99 + 28832112 ------GGUACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUCG------UGGGUUAACCUUUUU-- ------(((...........((((((((.....)))....)))))......((((((((.....))))))))......(((((((....)------.))))))))).....-- ( -27.80, z-score = -2.71, R) >droEre2.scaffold_4770 9778611 105 - 17746568 ------GGUACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUCGUGGGUUUGGGUUAACCUUUUU-- ------(((...........((((((((.....)))....)))))..((((((((((((.....)))))))).......((((((....).)))))))))...))).....-- ( -31.50, z-score = -2.53, R) >droVir3.scaffold_13047 4545951 94 + 19223366 -------GAACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUUUUGGCUGACAGCCAAAUUGUCGACAAUUUGUUGCUCUUAU----------UGUUGACUUUUUA-- -------.............((((((((.....)))....)))))..((((((.....))))))..(((((((((..(.....)..))----------)))))))......-- ( -25.50, z-score = -3.73, R) >droGri2.scaffold_14830 5291421 94 - 6267026 -------GAACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUUUUGGUCGACAGCCAAAUUGUCGACGAUUUGUUGCUCAUAU----------UGUUAACUUUUCA-- -------.............((((((((.....)))....)))))......((((((((.....))))))))((...(((((......----------.).))))...)).-- ( -20.60, z-score = -1.39, R) >droAna3.scaffold_13340 421425 105 - 23697760 GGAGCUCGUACUGACUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUCCCGAGCGACAGCCAAAUUGUCGACGAUUUGUUGCCGCUCUCC------UGUGUUAACCUUAUU-- (((((.((((.(((.....)))...(((.....)))...)))).))))).(((((.((((.((((((....)))))))))).)))))...------...............-- ( -29.20, z-score = -2.55, R) >droMoj3.scaffold_6540 16015705 94 + 34148556 -------GCACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUUUCGGCCGACAGCCAAAUUGUCGACGAUUUGUUGCUCUUAU----------UGUUGACUUUUGG-- -------...((((......((((((((.....)))....)))))...)))).......(((((..(((((((((..(.....)..))----------))))))).)))))-- ( -25.30, z-score = -2.42, R) >consensus ______GGUACUGAUUUUUUCACGCCCAUUAAUUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC________GGGUUAACCUUUUU__ ........(((.((((....((((((((.....)))....)))))........((((((.....))))))..)))).)))................................. (-14.45 = -14.34 + -0.11)

| Location | 10,245,403 – 10,245,493 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Shannon entropy | 0.31692 |
| G+C content | 0.42956 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.16 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 10245403 90 + 27905053 ACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGUACCGACG-------AUUGCAGUUUUGU---------------- ...(((((((((((.......)))).(((((...(((((....(((.....)))))))).....------.)))))...))))-------)))..........---------------- ( -24.20, z-score = -1.42, R) >droSim1.chr3R 16305116 90 - 27517382 ACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGUACCGACG-------AUUGCAGUUUUGU---------------- ...(((((((((((.......)))).(((((...(((((....(((.....)))))))).....------.)))))...))))-------)))..........---------------- ( -24.20, z-score = -1.42, R) >droSec1.super_0 9325184 90 - 21120651 ACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGUACCGACG-------AUUGCAGUUUUGU---------------- ...(((((((((((.......)))).(((((...(((((....(((.....)))))))).....------.)))))...))))-------)))..........---------------- ( -24.20, z-score = -1.42, R) >droYak2.chr3R 12636991 106 - 28832112 ACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGUACCGACG-------AUUGCAGUUUUGCUCUGUUGCAGUUUUGU ...(((((((((((.......)))).(((((...(((((....(((.....)))))))).....------.)))))...))))-------)))((((..((((......))))..)))) ( -31.20, z-score = -1.66, R) >droEre2.scaffold_4770 9778643 90 + 17746568 ACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGUACCGCCG-------AUUGCAGUUUUGU---------------- .......(((((((.......)))))))((....)).(((((((....((((.(((((((....------.)))....)))).-------))))..)))))))---------------- ( -24.10, z-score = -1.10, R) >dp4.chr2 16297528 94 + 30794189 ACAAAUCGUCGACAAUUUGGCUGUCGGCCAGAAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAG---UAGAGUAAAACGAUUGCAGUUUUGU---------------- ...((((((..((..((((((.....))))))..(((((....(((.....)))))))).....------.....---....))...))))))..........---------------- ( -22.10, z-score = -1.17, R) >droVir3.scaffold_13047 4545973 89 - 19223366 ACAAAUUGUCGACAAUUUGGCUGUCAGCCAAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGU-UCGUCG-------AUUGCAGUUUUGU---------------- (((((..((((((..((((((.....))))))..(((((....(((.....)))))))).....------......-..))))-------))......)))))---------------- ( -25.00, z-score = -2.38, R) >droGri2.scaffold_14830 5291443 90 + 6267026 ACAAAUCGUCGACAAUUUGGCUGUCGACCAAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGU-UCGACG-------AUUGCAGUUUUGUU--------------- ...((((((((((..((((((....).)))))..(((((....(((.....)))))))).....------....).-))))))-------)))...........--------------- ( -23.80, z-score = -1.84, R) >droAna3.scaffold_13340 421451 102 + 23697760 ACAAAUCGUCGACAAUUUGGCUGUCGCUCGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAGUCAGUACGAGCUCCGACGGGUC-CGACUUCAGCUUUGU---------------- .....(((((((((.......))))(((((.((.(((((....(((.....))))))))......))....)))))...)))))...-...............---------------- ( -25.20, z-score = 0.46, R) >droWil1.scaffold_181089 2320228 98 - 12369635 ACAAAUCGUCGACAAUUUGGCUGUCGGCCAGAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAC-----AACAAAGCCAACAAAAAAAAAUUGCAGUUUUGU---------------- .......(((((((.......)))))))......(((((....(((.....))))))))......-----.(((((((...(((.......)))..)))))))---------------- ( -21.10, z-score = -0.99, R) >droMoj3.scaffold_6540 16015727 91 - 34148556 ACAAAUCGUCGACAAUUUGGCUGUCGGCCGAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAA------AUCAGUG-CGACGAC-----GACGGAGUUUUGU---------------- .....((((((((..((((((.....))))))..(((((....(((.....)))))))).....------....)).-)))))).-----.............---------------- ( -25.50, z-score = -1.04, R) >consensus ACAAAUCGUCGACAAUUUGGCUGUCGACCGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAA______AUCAGUACCGACG_______AUUGCAGUUUUGU________________ (((((.((((((((.......)))))))......(((((....(((.....)))))))).....................................).)))))................ (-18.25 = -18.16 + -0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:59 2011