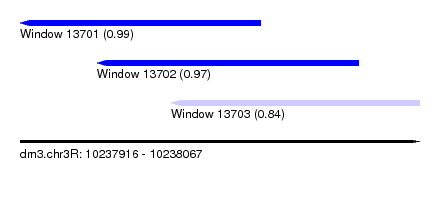
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,237,916 – 10,238,067 |
| Length | 151 |
| Max. P | 0.994879 |

| Location | 10,237,916 – 10,238,007 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 71.78 |
| Shannon entropy | 0.57220 |
| G+C content | 0.53751 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -14.64 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10237916 91 - 27905053 CCAAUUAAUG-----CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUGA--------GAUGAUCAGAUGCUCAGA-UGGCAUCGCCAUCGCCUU ....((((((-----.((-(((((((....))))))))).))))))...(((((...((((--------.....))))..)))..((-((((...))))))))... ( -37.40, z-score = -3.30, R) >droEre2.scaffold_4770 9771337 98 - 17746568 CCAAUUAAUG-----CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGCUCCGAUGCUCAGAUGUUCAGA-UGGCAUCGCCAUCGACU- ....((((((-----.((-(((((((....))))))))).))))))...............((((...((((((.((..(....)..-)))))))).))))....- ( -35.80, z-score = -2.17, R) >droYak2.chr3R 12629415 83 + 28832112 CCAAUUAAUG-----CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCAC----------------UCAGAUGCUCAGA-UGGCAUCGCCAUCGCCUU ....((((((-----.((-(((((((....))))))))).))))))...(((......----------------.....)))...((-((((...))))))..... ( -32.70, z-score = -3.07, R) >droSec1.super_0 9317703 91 + 21120651 CCAAUUAAUG-----CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCA--------GAUUAUCAGAUGCUCAGA-UGGCAUCGCCAUCGUCUU ....((((((-----.((-(((((((....))))))))).))))))...(((.........--------..........)))...((-((((...))))))..... ( -32.11, z-score = -2.80, R) >droSim1.chr3R 16297612 91 + 27517382 CCAAUUAAUG-----CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCA--------GAUGAUCAGAUGCUCAGA-UGGCAUCGCCAUCGCCUU ....((((((-----.((-(((((((....))))))))).))))))...(((..(((....--------..))).....)))...((-((((...))))))..... ( -34.10, z-score = -2.60, R) >dp4.chr2 16290206 78 - 30794189 CCAAUUAAUG-----CGUCCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUU----------------------GGGCUUUGGA-UGGAAUCGGAAUCGGAAU (((((...((-----(((((((((((....)))))))).........))))))))----------------------))..(((.((-(.........))).))). ( -26.90, z-score = -1.41, R) >droPer1.super_0 7714115 78 + 11822988 CCAAUUAAUG-----CGUCCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUU----------------------GGUCUUUGGA-UGGAAUCGGAAUCGGAAU ((..((((((-----.(.((((((((....))))))))).))))))..((...((----------------------.(((....))-).))..)).....))... ( -27.40, z-score = -1.75, R) >droAna3.scaffold_13340 415180 69 - 23697760 CCAAUUAAUG-----CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUC------------------------GGUCAGA-UGGCAUCGCCAU------ ....((((((-----.((-(((((((....))))))))).))))))..........------------------------......(-((((...)))))------ ( -26.80, z-score = -2.46, R) >droWil1.scaffold_181089 2313205 92 + 12369635 CCAAUUAAUGAUCGACGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGACCAGCA-----ACAACAUUUAGCAUCCAG--CAUC-UCGAUAUC----- ..........(((((.((-(((((((....))))))))).............((((.....))-----))........((.....)--)...-)))))...----- ( -25.60, z-score = -2.05, R) >droVir3.scaffold_13047 4539153 92 + 19223366 CCAAUUAAUG-----CGU-CGCCAGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGGCCCGUACGC-CAUGCCAUUGGGUAUUGGUAUCGGCAUCGACA------- ....((((((-----.((-(((((((....))))))))).))))))......(((((((.((((.(-.(((((....))))).))))).)))..)))).------- ( -40.10, z-score = -3.41, R) >droMoj3.scaffold_6540 16008824 89 + 34148556 CCAAUUAAUG-----CGU-CGCCAGCGAGAGUUGGCGGCUCAUUAGCACGCAGUUGGCUGAUAC------GCCAUUGGGCAUUGCUAUCGUCGACGACAUC----- ....((((((-----.((-(((((((....))))))))).))))))...(..((((((.((((.------(((....))).....))))))))))..)...----- ( -35.70, z-score = -2.78, R) >droGri2.scaffold_14830 5284299 91 - 6267026 CCAAUUAAUG-----CGU-CGCCAGCGAGAGUUGGCGGCUCAUUAACACGCAGUUUGUCCGAUC------GCCAUUGGGUAUUGGCAGCGGUAUCGGUAUCAU--- ((..((((((-----.((-(((((((....))))))))).))))))..(((....((.(((((.------(((....))))))))))))).....))......--- ( -35.90, z-score = -3.19, R) >consensus CCAAUUAAUG_____CGU_CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCA_________A__AUCAGGUGUUCAGA_UGGCAUCGCCAUCG____ ....((((((......((.(((((((....))))))))).))))))............................................................ (-14.64 = -14.38 + -0.26)
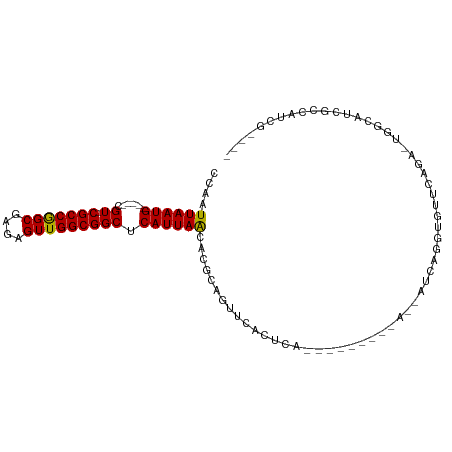

| Location | 10,237,945 – 10,238,044 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Shannon entropy | 0.50974 |
| G+C content | 0.56551 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -18.32 |
| Energy contribution | -18.07 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10237945 99 - 27905053 GCCCGGCCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUGAGAUGAUC------------ ....(((.............)))--...(((((((((((....((((((-.((-(((((((....))))))))).))))))...)).))))).......))))------------ ( -31.73, z-score = -1.03, R) >droEre2.scaffold_4770 9771372 100 - 17746568 UCCCUACCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGCUCC----------- ....................((.--..(((..(((((((....((((((-.((-(((((((....))))))))).))))))...)).))))).)))...))...----------- ( -32.50, z-score = -2.61, R) >droYak2.chr3R 12629436 100 + 28832112 GCCCGGCCAUGCCCAUAAACGCC-AAAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGCUC------------ ....(((...))).......((.-...(((..(((((((....((((((-.((-(((((((....))))))))).))))))...)).))))).)))...))..------------ ( -33.90, z-score = -1.67, R) >droSec1.super_0 9317732 99 + 21120651 GCCCGGCCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUUAUC------------ ....(((.............)))--.(((((((((((((....((((((-.((-(((((((....))))))))).))))))...)).)))....)))))))).------------ ( -34.22, z-score = -2.28, R) >droSim1.chr3R 16297641 99 + 27517382 GCCCGGCCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGAUC------------ ....(((.............)))--..(((..(((((((....((((((-.((-(((((((....))))))))).))))))...)).))))).))).......------------ ( -32.62, z-score = -1.45, R) >dp4.chr2 16290230 91 - 30794189 GCGUAGCCAUGCCCAUAAACGAC--AAUGAUCCGGACGCCCAAUUAAUG-CGUCCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGGGC--------------------- ..........(((((....((..--(((((.((((((((.........)-)))))((((((....)))))))).)))))....))....)))))--------------------- ( -36.40, z-score = -2.53, R) >droPer1.super_0 7714139 91 + 11822988 GCGUAGCCAUGCCCAUAAACGAC--AAUGAUCCGGACGCCCAAUUAAUG-CGUCCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGGUC--------------------- ((((.((...))...........--(((((.((((((((.........)-)))))((((((....)))))))).)))))...))))........--------------------- ( -32.70, z-score = -1.83, R) >droAna3.scaffold_13340 415195 91 - 23697760 UCGCGGCCAUGCCCGUAAACGAC--UAUGAUCCGGAUGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCGGUC-------------------- ..((((......)))).......--...((.((((((((....((((((-.((-(((((((....))))))))).))))))...))).)))))))-------------------- ( -35.10, z-score = -1.70, R) >droWil1.scaffold_181089 2313230 93 + 12369635 --------AUGCCCAUAAAAGAC-GAACGGGCCGACCAAUUAAUGAUCGACGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGACCAGCAACAAC------------ --------..((((.........-....)))).......(((((((.....((-(((((((....))))))))))))))))......((((.....))))...------------ ( -28.92, z-score = -2.06, R) >droVir3.scaffold_13047 4539172 109 + 19223366 --GUGUGGAUGCCCAUAAAAGAC-GUGGCGGAUGGCCGCCCAAUUAAUG-CGU-CGCCAGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGGCCCG-UACGCCAUGCCAUUGGGU --........(((((.....(.(-((((((...((((((....((((((-.((-(((((((....))))))))).))))))...))....))))..-..))))))))...))))) ( -49.20, z-score = -2.68, R) >droMoj3.scaffold_6540 16008845 107 + 34148556 ACCUGGCUAUGCCCAUAAAAGCCGCGACUGUUUGGCCGACCAAUUAAUG-CGU-CGCCAGCGAGAGUUGGCGGCUCAUUAGCACGCAGUUGGCUGAUACGCCAUUGGGC------ ..........(((((..........((((((((((....))))((((((-.((-(((((((....))))))))).))))))...))))))(((......)))..)))))------ ( -43.50, z-score = -2.10, R) >droGri2.scaffold_14830 5284322 104 - 6267026 --GUGUGGAUGCCCAUAAAAGAU-GUGGUGGUUGGCCGCCCAAUUAAUG-CGU-CGCCAGCGAGAGUUGGCGGCUCAUUAACACGCAGUUUGUCCG-AUCGC-----CAUUGGGU --........(((((........-((((((((((((.((....((((((-.((-(((((((....))))))))).))))))...)).....).)))-)))))-----)))))))) ( -43.60, z-score = -2.79, R) >consensus GCCCGGCCAUGCCCAUAAACGAC__AAUGAUCUGGACGCCCAAUUAAUG_CGU_CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCA_AUGAUC____________ .........(((.....................((....))..((((((.....(((((((....)))))))...))))))...)))............................ (-18.32 = -18.07 + -0.24)



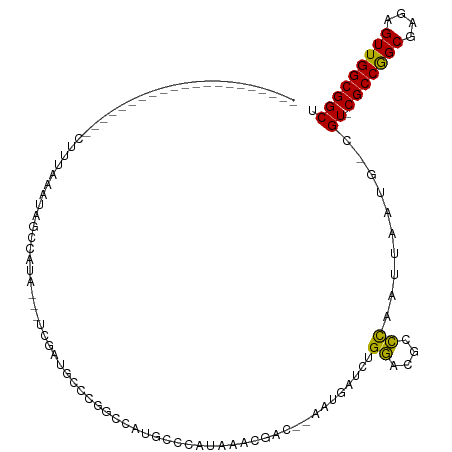
| Location | 10,237,973 – 10,238,067 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.53 |
| Shannon entropy | 0.51001 |
| G+C content | 0.55792 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -17.14 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10237973 94 - 27905053 ---------------------CUUUAAAUGGCCAUAUGUUCGGUGCCCGGCCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCU ---------------------......((((...((((..(((...)))..)))).))))....(((--.........(((((.........)-)))-)((((((....))))))))). ( -33.10, z-score = -0.76, R) >droEre2.scaffold_4770 9771401 94 - 17746568 ---------------------CUUUAAAUGGCCAUAUGCCUGCUUCCCUACCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCU ---------------------.......((((..((((...((...........)).))))...)))--).((((.(((....)))))))...-.((-(((((((....))))))))). ( -27.00, z-score = -0.55, R) >droYak2.chr3R 12629464 95 + 28832112 ---------------------CUUUAAAUGGCCACAUGUUCGGUGCCCGGCCAUGCCCAUAAACGCC-AAAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCU ---------------------......((((...((((..(((...)))..)))).))))....(((-..........(((((.........)-)))-)((((((....))))))))). ( -34.90, z-score = -1.25, R) >droSec1.super_0 9317760 94 + 21120651 ---------------------CUUUAAAUGGCCAUAUGUUCGGUGCCCGGCCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCU ---------------------......((((...((((..(((...)))..)))).))))....(((--.........(((((.........)-)))-)((((((....))))))))). ( -33.10, z-score = -0.76, R) >droSim1.chr3R 16297669 94 + 27517382 ---------------------CUUUAAAUGGCCAUAUGUUCGGUGCCCGGCCAUGCCCAUAAACGCC--AAUGAUCUGGACGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCU ---------------------......((((...((((..(((...)))..)))).))))....(((--.........(((((.........)-)))-)((((((....))))))))). ( -33.10, z-score = -0.76, R) >dp4.chr2 16290249 88 - 30794189 ---------------------CUUUAAAUAGCC-------CGAUGCGUAGCCAUGCCCAUAAACGAC--AAUGAUCCGGACGCCCAAUUAAUG-CGUCCGCCGGCGAGAGUUGGCGGCU ---------------------........((((-------..(((.(((....))).))).......--........((((((.........)-)))))((((((....)))))))))) ( -28.70, z-score = -1.54, R) >droPer1.super_0 7714158 88 + 11822988 ---------------------CUUUAAAUAGCC-------CGAUGCGUAGCCAUGCCCAUAAACGAC--AAUGAUCCGGACGCCCAAUUAAUG-CGUCCGCCGGCGAGAGUUGGCGGCU ---------------------........((((-------..(((.(((....))).))).......--........((((((.........)-)))))((((((....)))))))))) ( -28.70, z-score = -1.54, R) >droAna3.scaffold_13340 415215 111 - 23697760 AUAUAUAUGGCAGUGGCGCCUCUCGAAUCGGGCGCA----UGAUUCGCGGCCAUGCCCGUAAACGAC--UAUGAUCCGGAUGCCCAAUUAAUG-CGU-CGCCGGCGAGAGUUGGCGGCU ........(((....((((((........)))))).----..(((((.(..((((..((....))..--))))..))))))))).........-.((-(((((((....))))))))). ( -42.40, z-score = -0.38, R) >droWil1.scaffold_181089 2313258 85 + 12369635 ---------------------------CUAUAAAUA---GCCAUAAGUAG--AUGCCCAUAAAAGAC-GAACGGGCCGACCAAUUAAUGAUCGACGU-CGCCGGCGAGAGUUGGCGGCU ---------------------------........(---(((.......(--((((((.........-....))))(((.((.....)).)))..))-)((((((....)))))))))) ( -26.62, z-score = -2.26, R) >droVir3.scaffold_13047 4539211 84 + 19223366 ---------------------------CUAUACAUA---GUCAUGUGUGG--AUGCCCAUAAAAGAC-GUGGCGGAUGGCCGCCCAAUUAAUG-CGU-CGCCAGCGAGAGUUGGCGGCU ---------------------------((((((((.---...))))))))--..(((.......(((-(((((((....)))))........)-)))-)((((((....))))))))). ( -37.90, z-score = -3.89, R) >droMoj3.scaffold_6540 16008879 88 + 34148556 ---------------------------CGAGGCGCG--GCGCCUACCUGGCUAUGCCCAUAAAAGCCGCGACUGUUUGGCCGACCAAUUAAUG-CGU-CGCCAGCGAGAGUUGGCGGCU ---------------------------..((.((((--(((((.....)))(((....)))...)))))).))....(((((((((.....))-.))-)((((((....)))))))))) ( -38.80, z-score = -1.94, R) >droGri2.scaffold_14830 5284356 84 - 6267026 ---------------------------CUAUACAUA---GUCAUGUGUGG--AUGCCCAUAAAAGAU-GUGGUGGUUGGCCGCCCAAUUAAUG-CGU-CGCCAGCGAGAGUUGGCGGCU ---------------------------((((((((.---...))))))))--..(((.......(((-((..(((((((....)))))))..)-)))-)((((((....))))))))). ( -35.70, z-score = -3.54, R) >consensus _____________________CUUUAAAUAGCCAUA___UCGAUGCCCGGCCAUGCCCAUAAACGAC__AAUGAUCUGGACGCCCAAUUAAUG_CGU_CGCCGGCGAGAGUUGGCGGCU ......................................................(((....................((....))..............((((((....))))))))). (-17.14 = -16.73 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:55 2011