
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,226,925 – 10,227,027 |
| Length | 102 |
| Max. P | 0.771346 |

| Location | 10,226,925 – 10,227,027 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.44876 |
| G+C content | 0.36726 |
| Mean single sequence MFE | -17.74 |
| Consensus MFE | -10.08 |
| Energy contribution | -12.05 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
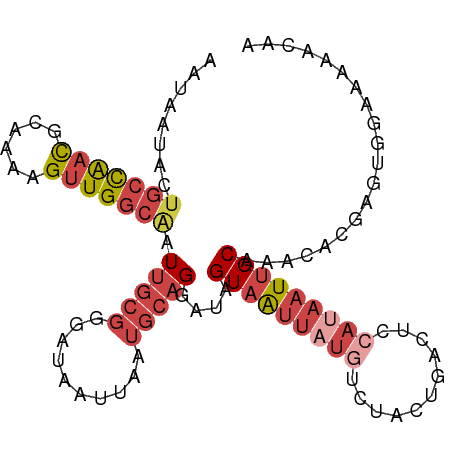
>dm3.chr3R 10226925 102 + 27905053 UUGUUUUUCCACUCGUGUUUGCAAUUAUGGAGUCAGUAGAUAUAAUUACUAUCCUGCAUUAAUUAUCCCGCAAUUGCCAACUUUUGCGUUGGUAGUAAUUUU .(((..(((((....((....))....)))))...(((((((.......))).))))............)))(((((((((......)))))))))...... ( -18.60, z-score = -0.93, R) >droSim1.chr3R 16288074 102 - 27517382 UUGUUUUUCCACUCGUGCUUGCAAUUAUGGAGUCAGUAGACAUAAUUACUAUCCUGCAUUAAUUAUCCCGCAAUUGCCAACUUUUACGUUGGCAGUAUUAUU .......((((....((....))....))))(((....))).............(((............)))(((((((((......)))))))))...... ( -20.20, z-score = -1.46, R) >droSec1.super_0 9306786 102 - 21120651 UUGUUUUUCCACUCGUGUUUGCAAUUAUGGAGUCAGUAGACAUAAUUACUAUCCUGCAUUAAUUAUCCCGCAAUUGCCAACUUUUGCGUUGGCUGUAUUAUU ........(((..((((((.((((((.(((.(((....)))(((((((...........))))))).))).)))))).)))....))).))).......... ( -17.30, z-score = -0.12, R) >droYak2.chr3R 12617959 102 - 28832112 CUGUUUUCCCACUCGUCUUUGCAAUUAUGCAGUCAGUAGACAUAAUUACUAUCCUGCAUUAAUUAUCCCGCAAUUGCCAACUUUUGCGUUGGCAGUAUUAUA ...................(((((((((((((..(((((......)))))...))))).))))).....)))(((((((((......)))))))))...... ( -22.80, z-score = -2.91, R) >droEre2.scaffold_4770 9760930 98 + 17746568 CUGUUUUUCCACUCGUGUUUGCAAUUUUGCA---AGUAGACAUAAUUACUAUCCUGCAUUAAUUAUCCCGCAAUUGCCAACUCU-GCGUUGGCAGUAUUAUU ..............(((.(((((....))))---)((((..............))))...........))).(((((((((...-..)))))))))...... ( -20.34, z-score = -1.99, R) >droVir3.scaffold_13047 4528147 88 - 19223366 UUAAUUAUCCAUGCGUA--UGCAA----AGAAUAGCUAGCGACAACAACUCUUCUAC--UAACAAGAGC---ACCGACAACUACAACAACAACAAUUCU--- ...........(((((.--.((..----......))..))).))....(((((....--....))))).---...........................--- ( -7.20, z-score = -0.14, R) >consensus UUGUUUUUCCACUCGUGUUUGCAAUUAUGGAGUCAGUAGACAUAAUUACUAUCCUGCAUUAAUUAUCCCGCAAUUGCCAACUUUUGCGUUGGCAGUAUUAUU ..............((((((((.............)))))))).............................(((((((((......)))))))))...... (-10.08 = -12.05 + 1.97)



| Location | 10,226,925 – 10,227,027 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.44876 |
| G+C content | 0.36726 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -11.61 |
| Energy contribution | -13.62 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10226925 102 - 27905053 AAAAUUACUACCAACGCAAAAGUUGGCAAUUGCGGGAUAAUUAAUGCAGGAUAGUAAUUAUAUCUACUGACUCCAUAAUUGCAAACACGAGUGGAAAAACAA .......((((((((......))))(((((((((..(....)..))).((((((((........)))))..)))..))))))........))))........ ( -19.60, z-score = -1.57, R) >droSim1.chr3R 16288074 102 + 27517382 AAUAAUACUGCCAACGUAAAAGUUGGCAAUUGCGGGAUAAUUAAUGCAGGAUAGUAAUUAUGUCUACUGACUCCAUAAUUGCAAGCACGAGUGGAAAAACAA .((((((((((((((......))))))..(((((..(....)..)))))...))).)))))(((....)))(((((...((....))...)))))....... ( -23.30, z-score = -1.60, R) >droSec1.super_0 9306786 102 + 21120651 AAUAAUACAGCCAACGCAAAAGUUGGCAAUUGCGGGAUAAUUAAUGCAGGAUAGUAAUUAUGUCUACUGACUCCAUAAUUGCAAACACGAGUGGAAAAACAA .....(((.((((((......))))))..(((((..(....)..)))))....(((((((((((....)))...))))))))........)))......... ( -22.40, z-score = -1.73, R) >droYak2.chr3R 12617959 102 + 28832112 UAUAAUACUGCCAACGCAAAAGUUGGCAAUUGCGGGAUAAUUAAUGCAGGAUAGUAAUUAUGUCUACUGACUGCAUAAUUGCAAAGACGAGUGGGAAAACAG .....((((((((((......))))))..((((((........((((((..(((((........))))).))))))..)))))).....))))......... ( -25.90, z-score = -2.55, R) >droEre2.scaffold_4770 9760930 98 - 17746568 AAUAAUACUGCCAACGC-AGAGUUGGCAAUUGCGGGAUAAUUAAUGCAGGAUAGUAAUUAUGUCUACU---UGCAAAAUUGCAAACACGAGUGGAAAAACAG .((((((((((((((..-...))))))..(((((..(....)..)))))...))).))))).((((((---((..............))))))))....... ( -23.04, z-score = -1.90, R) >droVir3.scaffold_13047 4528147 88 + 19223366 ---AGAAUUGUUGUUGUUGUAGUUGUCGGU---GCUCUUGUUA--GUAGAAGAGUUGUUGUCGCUAGCUAUUCU----UUGCA--UACGCAUGGAUAAUUAA ---................((((((((.((---((...(((..--...(((((((.((((....)))).)))))----)))))--...)))).)))))))). ( -21.40, z-score = -1.37, R) >consensus AAUAAUACUGCCAACGCAAAAGUUGGCAAUUGCGGGAUAAUUAAUGCAGGAUAGUAAUUAUGUCUACUGACUCCAUAAUUGCAAACACGAGUGGAAAAACAA ........(((((((......))))))).(((((..........)))))....(((((((((...........))))))))).................... (-11.61 = -13.62 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:52 2011