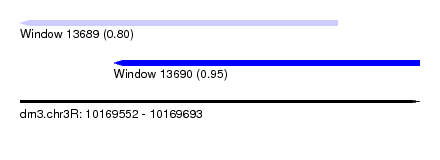
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,169,552 – 10,169,693 |
| Length | 141 |
| Max. P | 0.949917 |

| Location | 10,169,552 – 10,169,664 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.44 |
| Shannon entropy | 0.29813 |
| G+C content | 0.57440 |
| Mean single sequence MFE | -45.78 |
| Consensus MFE | -33.82 |
| Energy contribution | -33.75 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
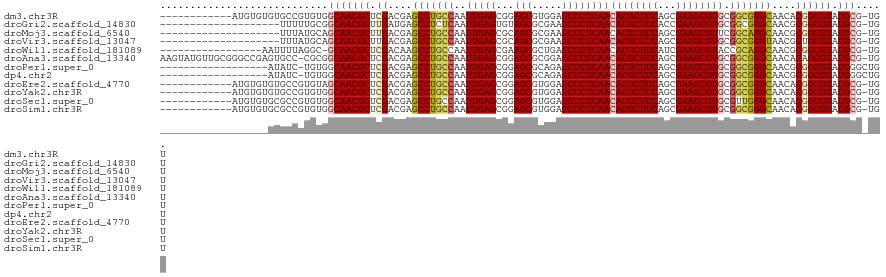
>dm3.chr3R 10169552 112 - 27905053 AGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGUUAUGCCAACUUAAAUGAUGCCCAUCG--GCAUCUU--- .(((((((........(((((.....)))))...((((((((...)))))))).))))))).....((((((....-...)))))).........((((((....)--)))))..--- ( -50.20, z-score = -3.05, R) >droGri2.scaffold_14830 5214844 117 - 6267026 AGCCUCUCAAUUGAGUGUAGCGCGAAGCUCUCAACACGCUUCACCGAAGCGUGCGGCGGGCAACGCGGCGUAUGCG-UGUUAUGCUAACUUAAAUGAUGCCCGACAGUGGCUCACACC ......(((.((((((.(((((((..((((.(..((((((((...))))))))..).))))..))).(((....))-).....)))))))))).))).((((....).)))....... ( -42.40, z-score = -1.14, R) >droMoj3.scaffold_6540 15935832 117 + 34148556 AGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUUCGGCAGGCAACGCGGCGUAUGCG-UGUUAUGCCAAAUUAAAUGAAGCUCAACCGUGGCUCAAAUU .(((((((....((((((..((((((((.........))))).)))..))))))))))))).....((((((....-...))))))........(((.((.((....))))))).... ( -46.10, z-score = -2.15, R) >droVir3.scaffold_13047 4455101 117 + 19223366 AGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGUAACGCUGCGUAUGCG-UGUUAUGCCAAAUUAAAUGACGCCCGACCGCGGCUCAAAUU .(((((((..(((((.((........)).)))))((((((((...)))))))).)))))))...((((((..((.(-((((((..........))))))).))..))))))....... ( -49.70, z-score = -2.29, R) >droWil1.scaffold_181089 2237066 112 + 12369635 AGCCUGCCAAUUGAGCGAAGCGCUGAGCUCUCAACACGCUUCAUCGAAGCGUACCGCAGGCAACGCGGCGUAUGCG-UGUUCUGCCAAAUUAAAUGAUGCCAGUCCGUUCAUA----- ...........((((((....((((.((..(((..(((((((...)))))))...(((((..(((((.....))))-)..))))).........))).)))))).))))))..----- ( -34.60, z-score = -0.27, R) >droAna3.scaffold_13340 354035 105 - 23697760 AGCCUGCCAAUUGAGCGGAGCGCGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACAGCGUAUGCG-UGUUCUGCCAACUCAAAUGGUGCCCAUCG------------ .((..((((.((((((((((((((..((((.(..((((((((...))))))))..).))))..)...((....)))-))))))))....)))).))))))......------------ ( -42.90, z-score = -1.14, R) >droPer1.super_0 7639129 115 + 11822988 AGCCUGCCAAUUGAGCGGAGCGCAGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACGCGGCGUAUGGGCUGUUCUGCCAACUUAAAUGAUGCCCAUCGCAGCAAAUU--- .(((((((........(((((.....)))))...((((((((...)))))))).)))))))...((.(((.((((((...((.............)).))))))))).)).....--- ( -51.62, z-score = -2.59, R) >dp4.chr2 16217621 115 - 30794189 AGCCUGCCAAUUGAGCGGAGCGCAGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACGCGGCGUAUGGGCUGUUCUGCCAACUUAAAUGAUGCCCAUCGCAGCAAAUU--- .(((((((........(((((.....)))))...((((((((...)))))))).)))))))...((.(((.((((((...((.............)).))))))))).)).....--- ( -51.62, z-score = -2.59, R) >droEre2.scaffold_4770 9703314 111 - 17746568 AGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGUUCUGCCAACUUAAAUGAUGCCCGUCG--GCAUCU---- .(((((((........(((((.....)))))...((((((((...)))))))).)))))))((((((.......))-))))..............((((((....)--))))).---- ( -47.40, z-score = -1.72, R) >droYak2.chr3R 12555154 112 + 28832112 AGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGUUCUGCCAACUUAAAUGAUGCCCAUCG--GCAUCUU--- .(((((((........(((((.....)))))...((((((((...)))))))).)))))))((((((.......))-))))..............((((((....)--)))))..--- ( -47.40, z-score = -2.08, R) >droSec1.super_0 9249873 112 + 21120651 AGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGUUGGGCAACACGGCGUAUGCG-UGUUCUGCCAACUUAAAUGACGCCCAACG--GCAUCUU--- ....((((......(((((((.....)))))....(((((((...)))))))))(((((((((((((.......))-)))).......(......)..))))))))--)))....--- ( -41.50, z-score = -0.80, R) >droSim1.chr3R 16230469 112 + 27517382 AGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGUUCUGCCAACUUAAAUGACGCCCAUCG--GCAUCUU--- ....((((..(((.((((((((((..((((.(..((((((((...))))))))..).))))..))).(((....))-))))))))))).....(((.....))).)--)))....--- ( -43.90, z-score = -1.01, R) >consensus AGCCUGCCAAUUGAGCGGAGCGCGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG_UGUUCUGCCAACUUAAAUGAUGCCCAUCG__GCAUCUU___ .(((((((..(((((...(((.....))))))))((((((((...)))))))).))))))).....(((((......((......)).........)))))................. (-33.82 = -33.75 + -0.07)
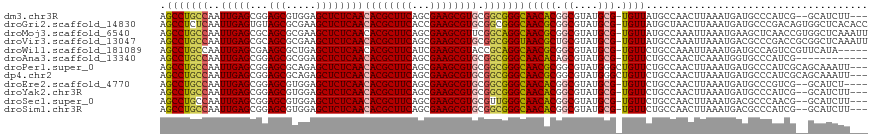
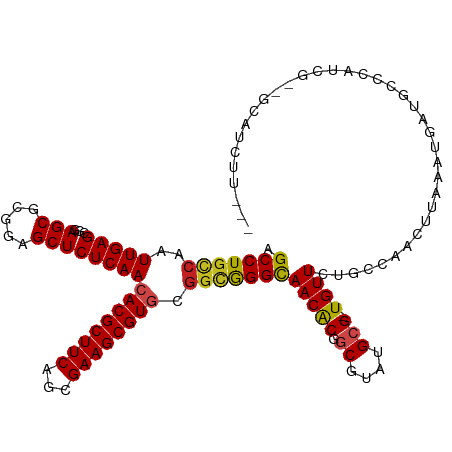
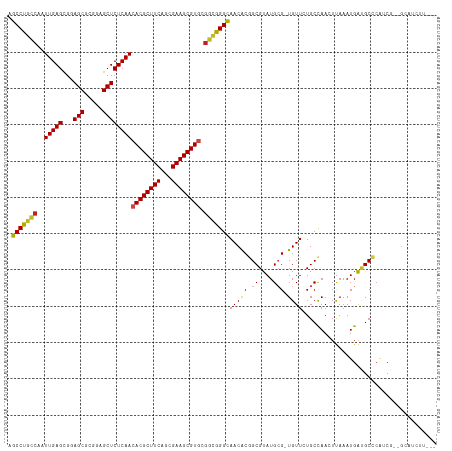
| Location | 10,169,585 – 10,169,693 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 84.29 |
| Shannon entropy | 0.31411 |
| G+C content | 0.61412 |
| Mean single sequence MFE | -49.63 |
| Consensus MFE | -38.83 |
| Energy contribution | -39.12 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.949917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
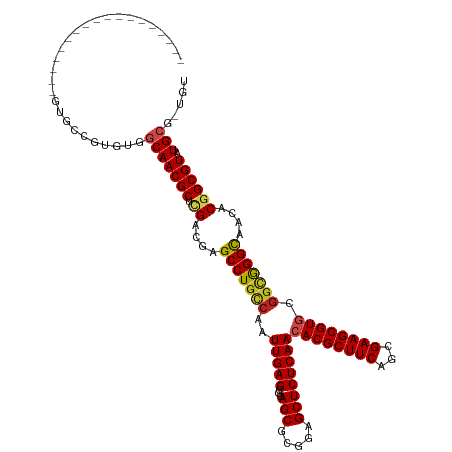
>dm3.chr3R 10169585 108 - 27905053 ------------AUGUGUGUGCCGUGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGU ------------((((((((((((((((....).........(((((((........(((((.....)))))...((((((((...)))))))).))))))).))))))))))))))-).. ( -53.60, z-score = -2.15, R) >droGri2.scaffold_14830 5214882 100 - 6267026 --------------------UUUUUGCGGCAACGCUUGAUGAGCCUCUCAAUUGAGUGUAGCGCGAAGCUCUCAACACGCUUCACCGAAGCGUGCGGCGGGCAACGCGGCGUAUGCG-UGU --------------------..((((((.(.(((((..(((((...))).))..))))).)))))))((((.(..((((((((...))))))))..).)))).(((((.....))))-).. ( -42.30, z-score = -1.51, R) >droMoj3.scaffold_6540 15935870 100 + 34148556 --------------------UUUAUGCAGCAACGCUUGACGAGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUUCGGCAGGCAACGCGGCGUAUGCG-UGU --------------------..((((((...(((((.(.((.(((((((....((((((..((((((((.........))))).)))..)))))))))))))..)))))))).))))-)). ( -46.90, z-score = -2.57, R) >droVir3.scaffold_13047 4455139 100 + 19223366 --------------------UUUAUGCAGCAACGCUUGACGAGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGUAACGCUGCGUAUGCG-UGU --------------------..((((((((..((.....)).(((((((..(((((.((........)).)))))((((((((...)))))))).)))))))...))))))))....-... ( -44.00, z-score = -1.27, R) >droWil1.scaffold_181089 2237099 102 + 12369635 -----------------AAUUUUAGGC-GCAACGCUCGACAAGCCUGCCAAUUGAGCGAAGCGCUGAGCUCUCAACACGCUUCAUCGAAGCGUACCGCAGGCAACGCGGCGUAUGCG-UGU -----------------........((-(((((((.((....((((((...(((((...(((.....)))))))).(((((((...)))))))...))))))....)))))).))))-).. ( -42.00, z-score = -2.21, R) >droAna3.scaffold_13340 354061 119 - 23697760 AAGUAUGUUGCGGGCCGAGUGCC-CGCGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACAGCGUAUGCG-UGU ..(((((((((((((.....)))-)))))).(((((......(((((((........(((((.....)))))...((((((((...)))))))).))))))).....))))))))).-... ( -57.70, z-score = -1.71, R) >droPer1.super_0 7639164 102 + 11822988 ------------------AUAUC-UGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCAGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACGCGGCGUAUGGGCUGU ------------------.....-.(..((.((((.((.((.(((((((........(((((.....)))))...((((((((...)))))))).)))))))..))))))))....))..) ( -44.20, z-score = -0.94, R) >dp4.chr2 16217656 102 - 30794189 ------------------AUAUC-UGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCAGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACGCGGCGUAUGGGCUGU ------------------.....-.(..((.((((.((.((.(((((((........(((((.....)))))...((((((((...)))))))).)))))))..))))))))....))..) ( -44.20, z-score = -0.94, R) >droEre2.scaffold_4770 9703346 108 - 17746568 ------------AUGUGUGUGCCGUGUAGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGU ------------(((((((((((((((.((...((((...))))(((((........(((((.....)))))...((((((((...)))))))).))))))).))))))))))))))-).. ( -52.40, z-score = -2.12, R) >droYak2.chr3R 12555187 108 + 28832112 ------------AUGUGUGUGCCGUGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGU ------------((((((((((((((((....).........(((((((........(((((.....)))))...((((((((...)))))))).))))))).))))))))))))))-).. ( -53.60, z-score = -2.15, R) >droSec1.super_0 9249906 108 + 21120651 ------------AUGUGUGCGCCGUGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGUUGGGCAACACGGCGUAUGCG-UGU ------------(((((((((((((((.((...((((.(((..((.(((....).))))..))).))))..((((((((((((...)))))))..))))))).))))))))))))))-).. ( -56.30, z-score = -3.11, R) >droSim1.chr3R 16230502 108 + 27517382 ------------AUGUGUGCGCCGUGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG-UGU ------------((((((((((((((((....).........(((((((........(((((.....)))))...((((((((...)))))))).))))))).))))))))))))))-).. ( -58.40, z-score = -3.27, R) >consensus __________________GUGCCGUGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCG_UGU ............................(((((((.((....(((((((..(((((...(((.....))))))))((((((((...)))))))).)))))))....)))))).)))..... (-38.83 = -39.12 + 0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:45 2011