
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,110,207 – 10,110,321 |
| Length | 114 |
| Max. P | 0.716378 |

| Location | 10,110,207 – 10,110,321 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.58717 |
| G+C content | 0.40367 |
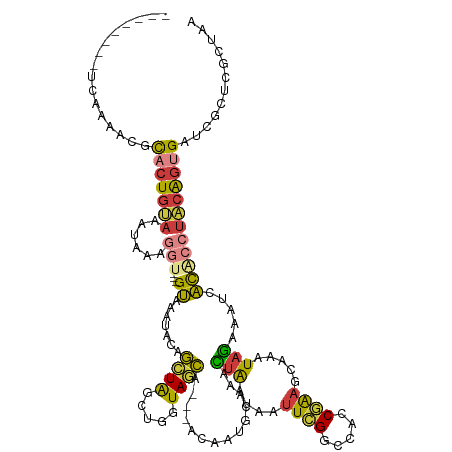
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -11.11 |
| Energy contribution | -10.61 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
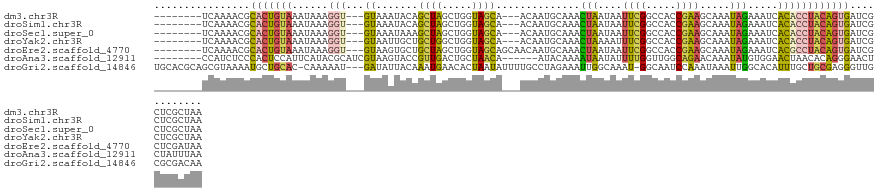
>dm3.chr3R 10110207 114 + 27905053 --------UCAAAACGCACUGUAAAUAAAGGU---GUAAAUACAGCUAGCUGGUAGCA---ACAAUGCAAACUAAUAAUUCGGCCACCGAAGCAAAUAGAAAUCACACCUACAGUGAUCGCUCGCUAA --------........((((((......((((---((.....(((....)))...(((---....)))...(((....(((((...))))).....))).....))))))))))))............ ( -24.00, z-score = -1.35, R) >droSim1.chr3R 16171868 114 - 27517382 --------UCAAAACGCACUGUAAAUAAAGGU---GUAAAUACAGCUAGCUGGUAGCA---ACAAUGCAAACUAAUAAUUCGGCCACCGAAGCAAAUAGAAAUCACACCUACAGUGAUCGCUCGCUAA --------........((((((......((((---((.....(((....)))...(((---....)))...(((....(((((...))))).....))).....))))))))))))............ ( -24.00, z-score = -1.35, R) >droSec1.super_0 9190904 114 - 21120651 --------UCAAAACGCACUGUAAAUAAAGGU---GUAAAUAAAGCUAGCUGGUAGCA---ACAAUGCAAACUAAUAAUUCGGCCACCGAAGCAAAUAGAAAUCACACCUACAGUGAUCGCUCGCUAA --------........((((((......((((---((.......((((.....)))).---..........(((....(((((...))))).....))).....))))))))))))............ ( -22.80, z-score = -1.08, R) >droYak2.chr3R 12494700 114 - 28832112 --------UCAAAACGCACUGUAAAUAAAGGU---GUAAUUGCUGCUGGCUGGUAGCA---ACAAUGCAAACUAAAAUUUCGGCCACCGAAGCAAAUAGAAAUCACACCUACAGUGAUCGCUCGCUAA --------........((((((......((((---((..((((((((....)))))))---).........(((...((((((...))))))....))).....))))))))))))............ ( -28.70, z-score = -1.81, R) >droEre2.scaffold_4770 9643535 117 + 17746568 --------UCAAAACGCACUGUAAAUAAAGGU---GUAAGUGCUGCUAGCUGGUAGCAGCAACAAUGCAAACUAAUAAUUCGGCCACCGAAGCAAAUAGAAAUCACGCCUACAGUGAUCGCUCGAUAA --------........((((((......((((---((...((((((((.....))))))))..........(((....(((((...))))).....))).....))))))))))))(((....))).. ( -31.20, z-score = -1.96, R) >droAna3.scaffold_12911 2831599 114 - 5364042 --------CCAUCUCCCACUCCAUUCAUACGCAUCGUAAGUACCGUUGACUGCUAACA------AUACAAAAUAAUAUUUUGGUUGGCAGAACAAAUAUGUGGAACUAACACAGGGAACUCUAUUUAA --------.....((((.......(((.(((...(....)...))))))((((((((.------...((((((...))))))))))))))........((((.......))))))))........... ( -25.00, z-score = -2.28, R) >droGri2.scaffold_14846 190805 123 + 266023 UGCACGCAGCGUAAAAUGCUGCAC-CAAAAAU---GAUAUUACAAAUGAACACUAAUAUUUUGCCUAGAAAUUGGCAAAU-GGCAAUCCAAAUAAAUUGGCACAUUUGCUGCGAGGGUUGCGCGACAA .((..(((((((...)))))))..-.......---........................((((((........)))))).-.(((((((.........((((....))))....)))))))))..... ( -29.52, z-score = -0.39, R) >consensus ________UCAAAACGCACUGUAAAUAAAGGU___GUAAAUACAGCUAGCUGGUAGCA___ACAAUGCAAACUAAUAAUUCGGCCACCGAAGCAAAUAGAAAUCACACCUACAGUGAUCGCUCGCUAA ................(((((((.....................((((.....))))..............(((....((((.....)))).....)))..........)))))))............ (-11.11 = -10.61 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:14:37 2011